Transcription factor Gfi-1 induced by G-CSF is a negative regulator of CXCR4 in myeloid cells
- PMID: 17596540
- PMCID: PMC1988943
- DOI: 10.1182/blood-2007-03-081448
Transcription factor Gfi-1 induced by G-CSF is a negative regulator of CXCR4 in myeloid cells
Abstract
The mechanisms underlying granulocyte-colony stimulating factor (G-CSF)-induced mobilization of granulocytic lineage cells from the bone marrow to the peripheral blood remain elusive. We provide evidence that the transcriptional repressor growth factor independence-1 (Gfi-1) is involved in G-CSF-induced mobilization of granulocytic lineage cells from the bone marrow to the peripheral blood. We show that in vitro and in vivo G-CSF promotes expression of Gfi-1 and down-regulates expression of CXCR4, a chemokine receptor essential for the retention of hematopoietic stem cells and granulocytic cells in the bone marrow. Gfi-1 binds to DNA sequences upstream of the CXCR4 gene and represses CXCR4 expression in myeloid lineage cells. As a consequence, myeloid cell responses to the CXCR4 unique ligand SDF-1 are reduced. Thus, Gfi-1 not only regulates hematopoietic stem cell function and myeloid cell development but also probably promotes the release of granulocytic lineage cells from the bone marrow to the peripheral blood by reducing CXCR4 expression and function.
Figures
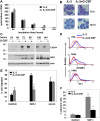

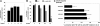




References
-
- Lieschke GJ, Grail D, Hodgson G, et al. Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor cell deficiency, and impaired neutrophil mobilization. Blood. 1994;84:1737–1746. - PubMed
-
- Liu F, Wu HY, Wesselschmidt R, Kornaga T, Link DC. Impaired production and increased apoptosis of neutrophils in granulocyte colony-stimulating factor receptor-deficient mice. Immunity. 1996;5:491–501. - PubMed
-
- Druhan LJ, Ai J, Massullo P, Kindwall-Keller T, Ranalli MA, Avalos BR. Novel mechanism of G-CSF refractoriness in patients with severe congenital neutropenia. Blood. 2005;105:584–591. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

