Modulating mtDNA heteroplasmy by mitochondria-targeted restriction endonucleases in a 'differential multiple cleavage-site' model
- PMID: 17597792
- PMCID: PMC2771437
- DOI: 10.1038/sj.gt.3302981
Modulating mtDNA heteroplasmy by mitochondria-targeted restriction endonucleases in a 'differential multiple cleavage-site' model
Abstract
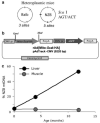
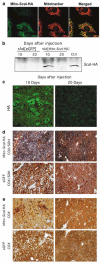
The ability to manipulate mitochondrial DNA (mtDNA) heteroplasmy would provide a powerful tool to treat mitochondrial diseases. Recent studies showed that mitochondria-targeted restriction endonucleases can modify mtDNA heteroplasmy in a predictable and efficient manner if it recognizes a single site in the mutant mtDNA. However, the applicability of such model is limited to mutations that create a novel cleavage site, not present in the wild-type mtDNA. We attempted to extend this approach to a 'differential multiple cleavage site' model, where an mtDNA mutation creates an extra restriction site to the ones normally present in the wild-type mtDNA. Taking advantage of a heteroplasmic mouse model harboring two haplotypes of mtDNA (NZB/BALB) and using adenovirus as a gene vector, we delivered a mitochondria-targeted Scal restriction endonuclease to different mouse tissues. Scal recognizes five sites in the NZB mtDNA but only three in BALB mtDNA. Our results showed that changes in mtDNA heteroplasmy were obtained by the expression of mitochondria-targeted ScaI in both liver, after intravenous injection, and in skeletal muscle, after intramuscular injection. Although mtDNA depletion was an undesirable side effect, our data suggest that under a regulated expression system, mtDNA depletion could be minimized and restriction endonucleases recognizing multiple sites could have a potential for therapeutic use.
Figures




References
-
- Schaefer AM, Taylor RW, Turnbull DM, Chinnery PF. The epidemiology of mitochondrial disorders - past, present and future. Biochim Biophys Acta. 2004;1659:115–120. - PubMed
-
- Chinnery PF, Johnson MA, Wardell TM, Singh-Kler R, Hayes C, Brown DT, et al. The epidemiology of pathogenic mitochondrial DNA mutations. Ann Neurol. 2000;48:188–193. - PubMed
-
- Chomyn A, Martinuzzi A, Yoneda M, Daga A, Hurko O, Johns D, et al. MELAS mutation in mtDNA binding site for transcription termination factor causes defects in protein synthesis and in respiration but no change in levels of upstream and downstream mature transcripts. Proc Natl Acad Sci USA. 1992;89:4221–4225. - PMC - PubMed
-
- Hanna MG, Nelson IP, Morgan-Hughes JA, Harding AE. Impaired mitochondrial translation in human myoblasts har-bouring the mitochondrial DNA tRNA lysine 8344 A→G (MERRF) mutation: relationship to proportion of mutant mitochondrial DNA. J Neurol Sci. 1995;130:154–160. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

