Integrative analysis of the mouse embryonic transcriptome
- PMID: 17597930
- PMCID: PMC1896055
- DOI: 10.6026/97320630001406
Integrative analysis of the mouse embryonic transcriptome
Abstract
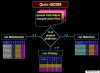
Monitoring global gene expression provides insight into how genes and regulatory signals work together to guide embryo development. The fields of developmental biology and teratology are now confronted with the need for automated access to a reference library of gene-expression signatures that benchmark programmed (genetic) and adaptive (environmental) regulation of the embryonic transcriptome. Such a library must be constructed from highly-distributed microarray data. Birth Defects Systems Manager (BDSM), an open access knowledge management system, provides custom software to mine public microarray data focused on developmental health and disease. The present study describes tools for seamless data integration in the BDSM library (MetaSample, MetaChip, CIAeasy) using the QueryBDSM module. A field test of the prototype was run using published microarray data series derived from a variety of laboratories, experiments, microarray platforms, organ systems, and developmental stages. The datasets focused on several developing systems in the mouse embryo, including preimplantation stages, heart and nerve development, testis and ovary development, and craniofacial development. Using BDSM data integration tools, a gene-expression signature for 346 genes was resolved that accurately classified samples by organ system and developmental sequence. The module builds a potential for the BDSM approach to decipher a large number developmental processes through comparative bioinformatics analysis of embryological systems at-risk for specific defects, using multiple scenarios to define the range of probabilities leading from molecular phenotype to clinical phenotype. We conclude that an integrative analysis of global gene-expression of the developing embryo can form the foundation for constructing a reference library of signaling pathways and networks for normal and abnormal regulation of the embryonic transcriptome. These tools are available free of charge from the web-site http://systemsanalysis.louisville.edu requiring only a short registration process.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
