Relationship between insertion/deletion (indel) frequency of proteins and essentiality
- PMID: 17598914
- PMCID: PMC1925122
- DOI: 10.1186/1471-2105-8-227
Relationship between insertion/deletion (indel) frequency of proteins and essentiality
Abstract
Background: In a previous study, we demonstrated that some essential proteins from pathogenic organisms contained sizable insertions/deletions (indels) when aligned to human proteins of high sequence similarity. Such indels may provide sufficient spatial differences between the pathogenic protein and human proteins to allow for selective targeting. In one example, an indel difference was targeted via large scale in-silico screening. This resulted in selective antibodies and small compounds which were capable of binding to the deletion-bearing essential pathogen protein without any cross-reactivity to the highly similar human protein. The objective of the current study was to investigate whether indels were found more frequently in essential than non-essential proteins.
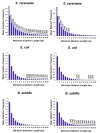
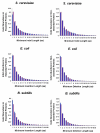
Results: We have investigated three species, Bacillus subtilis, Escherichia coli, and Saccharomyces cerevisiae, for which high-quality protein essentiality data is available. Using these data, we demonstrated with t-test calculations that the mean indel frequencies in essential proteins were greater than that of non-essential proteins in the three proteomes. The abundance of indels in both types of proteins was also shown to be accurately modeled by the Weibull distribution. However, Receiver Operator Characteristic (ROC) curves showed that indel frequencies alone could not be used as a marker to accurately discriminate between essential and non-essential proteins in the three proteomes. Finally, we analyzed the protein interaction data available for S. cerevisiae and observed that indel-bearing proteins were involved in more interactions and had greater betweenness values within Protein Interaction Networks (PINs).
Conclusion: Overall, our findings demonstrated that indels were not randomly distributed across the studied proteomes and were likely to occur more often in essential proteins and those that were highly connected, indicating a possible role of sequence insertions and deletions in the regulation and modification of protein-protein interactions. Such observations will provide new insights into indel-based drug design using bioinformatics and cheminformatics tools.
Figures


References
-
- Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, Fleischmann RD, Bult CJ, Kerlavage AR, Sutton G, Kelley JM, Fritchman RD, Weidman JF, Small KV, Sandusky M, Fuhrmann J, Nguyen D, Utterback TR, Saudek DM, Phillips CA, Merrick JM, Tomb JF, Dougherty BA, Bott KF, Hu PC, Lucier TS, Peterson SN, Smith HO, Hutchison CA, 3, Venter JC. The minimum gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

