BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges
- PMID: 17599064
- PMCID: PMC1933408
- DOI: 10.1038/sj.emboj.7601777
BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges
Abstract
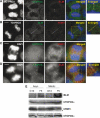
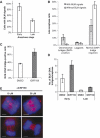
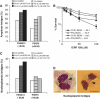
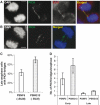
Mutations in BLM cause Bloom's syndrome, a disorder associated with cancer predisposition and chromosomal instability. We investigated whether BLM plays a role in ensuring the faithful chromosome segregation in human cells. We show that BLM-defective cells display a higher frequency of anaphase bridges and lagging chromatin than do isogenic corrected derivatives that eptopically express the BLM protein. In normal cells undergoing mitosis, BLM protein localizes to anaphase bridges, where it colocalizes with its cellular partners, topoisomerase IIIalpha and hRMI1 (BLAP75). Using BLM staining as a marker, we have identified a class of ultrafine DNA bridges in anaphase that are surprisingly prevalent in the anaphase population of normal human cells. These so-called BLM-DNA bridges, which also stain for the PICH protein, frequently link centromeric loci, and are present at an elevated frequency in cells lacking BLM. On the basis of these results, we propose that sister-chromatid disjunction is often incomplete in human cells even after the onset of anaphase. We present a model for the action of BLM in ensuring complete sister chromatid decatenation in anaphase.
Figures





References
-
- Baumann C, Korner R, Hofmann K, Nigg EA (2007) PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell 128: 101–114 - PubMed
-
- Beamish H, Kedar P, Kaneko H, Chen P, Fukao T, Peng C, Beresten S, Gueven N, Purdie D, Lees-Miller S, Ellis N, Kondo N, Lavin MF (2002) Functional link between BLM defective in Bloom's syndrome and the ataxia-telangiectasia-mutated protein, ATM. J Biol Chem 277: 30515–30523 - PubMed
-
- Bennink ML, Leuba SH, Leno GH, Zlatanova J, de Grooth BG, Greve J (2001) Unfolding individual nucleosomes by stretching single chromatin fibers with optical tweezers. Nat Struct Biol 8: 606–610 - PubMed
-
- Branzei D, Sollier J, Liberi G, Zhao X, Maeda D, Seki M, Enomoto T, Ohta K, Foiani M (2006) Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127: 509–522 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

