Transcription termination defective mutants of Rho: role of different functions of Rho in releasing RNA from the elongation complex
- PMID: 17599352
- PMCID: PMC1950744
- DOI: 10.1016/j.jmb.2007.06.013
Transcription termination defective mutants of Rho: role of different functions of Rho in releasing RNA from the elongation complex
Abstract
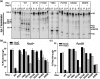
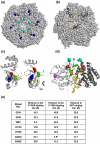
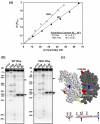
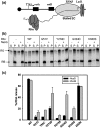
The transcription termination factor Rho of Escherichia coli is a RNA binding protein which can translocate along the RNA and unwind the RNA:DNA hybrid using the RNA-dependent ATPase activity. In order to investigate the involvement of each of these functions in releasing RNA from the elongation complex, we have isolated different termination defective mutants of Rho by random mutagenesis, characterized them for their different functions and established the structure-function correlations from the available structural data of Rho. These mutations are located within the two domains; the N-terminal RNA binding domain (G51V, G53V, and Y80C) and in the C-terminal ATP binding domain (Y274D, P279S, P279L, G324D, N340S, I382N) including the two important structural elements, the Q-loop (P279S, P279L) and R-loop (G324D). Termination defects of the mutants in primary RNA binding domain and Q-loop could not be restored under any conditions that we tested and these were also defective for most of the other functions of Rho. The termination defects of the mutants (Y274D, G324D and N340S), which were mainly defective for secondary RNA binding and likely defective for translocase activity, could be restored under relaxed in vitro conditions. We also show that a mutation in a primary RNA binding domain (Y80C) can cause a defect in ATP binding and induce distinct conformational changes in the distal C-terminal domain, and these allosteric effects are not predictable from the crystal structure. We conclude that the interactions in the primary RNA binding domain and in the Q-loop are mandatory for RNA release to occur and propose that the interactions in the primary RNA binding modulate most of the other functions of Rho allosterically. The rate of ATP hydrolysis regulates the processivity of translocation along the RNA and is directly correlated with the efficiency of RNA release. NusG improves the speed of RNA release and is not involved in any other step.
Figures


References
-
- Wilson K.S., von Hippel P.H. Stability of Escherichia coli transcription complexes near an intrinsic terminator. J. Mol. Biol. 1994;244:36–51. - PubMed
-
- Yarnell W.S., Roberts J.W. Mechanism of intrinsic transcription termination and antitermination. Science. 1999;284:611–615. - PubMed
-
- Richardson J.P. Transcription termination. Crit. Rev. Biochem. Mol. Biol. 1993;28:1–30. - PubMed
-
- Uptain S.M., Kane C.M., Chamberlin M.J. Basic mechanisms of transcript elongation and its regulation. Annu. Rev. Biochem. 1997;66:117–172. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

