DNA repair glycosylases with a [4Fe-4S] cluster: a redox cofactor for DNA-mediated charge transport?
- PMID: 17599416
- PMCID: PMC2094209
- DOI: 10.1016/j.jinorgbio.2007.05.001
DNA repair glycosylases with a [4Fe-4S] cluster: a redox cofactor for DNA-mediated charge transport?
Abstract
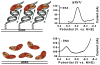
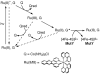
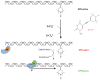
The [4Fe-4S] cluster is ubiquitous to a class of base excision repair enzymes in organisms ranging from bacteria to man and was first considered as a structural element, owing to its redox stability under physiological conditions. When studied bound to DNA, two of these repair proteins (MutY and Endonuclease III from Escherichia coli) display DNA-dependent reversible electron transfer with characteristics typical of high potential iron proteins. These results have inspired a reexamination of the role of the [4Fe-4S] cluster in this class of enzymes. Might the [4Fe-4S] cluster be used as a redox cofactor to search for damaged sites using DNA-mediated charge transport, a process well known to be highly sensitive to lesions and mismatched bases? Described here are experiments demonstrating the utility of DNA-mediated charge transport in characterizing these DNA-binding metalloproteins, as well as efforts to elucidate this new function for DNA as an electronic signaling medium among the proteins.
Figures



References
-
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T, editors. DNA Repair and Mutagenesis. ASM Press; Washington DC: 2006.
-
- David SS, Williams SD. Chem Rev. 1998;98:1221–1262. - PubMed
-
- Francis AW, David SS. Biochemistry. 2003;42:801–810. - PubMed
-
- Halford SE, Szczelkun MD. Eur Biophys J. 2002;31:257–267. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

