Characterization of mutation spectra with ultra-deep pyrosequencing: application to HIV-1 drug resistance
- PMID: 17600086
- PMCID: PMC1933516
- DOI: 10.1101/gr.6468307
Characterization of mutation spectra with ultra-deep pyrosequencing: application to HIV-1 drug resistance
Abstract
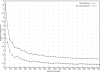
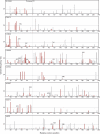
The detection of mutant spectra within a population of microorganisms is critical for the management of drug-resistant infections. We performed ultra-deep pyrosequencing to detect minor sequence variants in HIV-1 protease and reverse transcriptase (RT) genes from clinical plasma samples. We estimated empirical error rates from four HIV-1 plasmid clones and used them to develop a statistical approach to distinguish authentic minor variants from sequencing errors in eight clinical samples. Ultra-deep pyrosequencing detected an average of 58 variants per sample compared with an average of eight variants per sample detected by conventional direct-PCR dideoxynucleotide sequencing. In the clinical sample with the largest number of minor sequence variants, all 60 variants present in > or =3% of genomes and 20 of 35 variants present in <3% of genomes were confirmed by limiting dilution sequencing. With appropriate analysis, ultra-deep pyrosequencing is a promising method for characterizing genetic diversity and detecting minor yet clinically relevant variants in biological samples with complex genetic populations.
Figures
References
-
- Cai F., Chen H., Hicks C.B., Bartlett J.A., Zhu J., Gao F., Chen H., Hicks C.B., Bartlett J.A., Zhu J., Gao F., Hicks C.B., Bartlett J.A., Zhu J., Gao F., Bartlett J.A., Zhu J., Gao F., Zhu J., Gao F., Gao F. Detection of minor drug-resistant populations by parallel allele-specific sequencing. Nat. Methods. 2007;4:123–125. - PubMed
-
- Coffin J.M. HIV population dynamics in vivo: Implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. - PubMed
-
- Edelstein R.E., Nickerson D.A., Tobe V.O., Manns-Arcuino L.A., Frenkel L.M., Nickerson D.A., Tobe V.O., Manns-Arcuino L.A., Frenkel L.M., Tobe V.O., Manns-Arcuino L.A., Frenkel L.M., Manns-Arcuino L.A., Frenkel L.M., Frenkel L.M. Oligonucleotide ligation assay for detecting mutations in the human immunodeficiency virus type 1 pol gene that are associated with resistance to zidovudine, didanosine, and lamivudine. J. Clin. Microbiol. 1998;36:569–572. - PMC - PubMed
-
- Ellis G.M., Mahalanabis M., Beck I.A., Pepper G., Wright A., Hamilton S., Holte S., Naugler W.E., Pawluk D.M., Li C.C., Mahalanabis M., Beck I.A., Pepper G., Wright A., Hamilton S., Holte S., Naugler W.E., Pawluk D.M., Li C.C., Beck I.A., Pepper G., Wright A., Hamilton S., Holte S., Naugler W.E., Pawluk D.M., Li C.C., Pepper G., Wright A., Hamilton S., Holte S., Naugler W.E., Pawluk D.M., Li C.C., Wright A., Hamilton S., Holte S., Naugler W.E., Pawluk D.M., Li C.C., Hamilton S., Holte S., Naugler W.E., Pawluk D.M., Li C.C., Holte S., Naugler W.E., Pawluk D.M., Li C.C., Naugler W.E., Pawluk D.M., Li C.C., Pawluk D.M., Li C.C., Li C.C., et al. Comparison of oligonucleotide ligation assay and consensus sequencing for detection of drug-resistant mutants of human immunodeficiency virus type 1 in peripheral blood mononuclear cells and plasma. J. Clin. Microbiol. 2004;42:3670–3674. - PMC - PubMed
-
- Flys T., Nissley D.V., Claasen C.W., Jones D., Shi C., Guay L.A., Musoke P., Mmiro F., Strathern J.N., Jackson J.B., Nissley D.V., Claasen C.W., Jones D., Shi C., Guay L.A., Musoke P., Mmiro F., Strathern J.N., Jackson J.B., Claasen C.W., Jones D., Shi C., Guay L.A., Musoke P., Mmiro F., Strathern J.N., Jackson J.B., Jones D., Shi C., Guay L.A., Musoke P., Mmiro F., Strathern J.N., Jackson J.B., Shi C., Guay L.A., Musoke P., Mmiro F., Strathern J.N., Jackson J.B., Guay L.A., Musoke P., Mmiro F., Strathern J.N., Jackson J.B., Musoke P., Mmiro F., Strathern J.N., Jackson J.B., Mmiro F., Strathern J.N., Jackson J.B., Strathern J.N., Jackson J.B., Jackson J.B., et al. Sensitive drug-resistance assays reveal long-term persistence of HIV-1 variants with the K103N nevirapine (NVP) resistance mutation in some women and infants after the administration of single-dose NVP: HIVNET 012. J. Infect. Dis. 2005;192:24–29. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
