Evaluating the state-of-the-art in automatic de-identification
- PMID: 17600094
- PMCID: PMC1975792
- DOI: 10.1197/jamia.M2444
Evaluating the state-of-the-art in automatic de-identification
Abstract
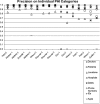
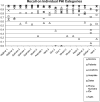
To facilitate and survey studies in automatic de-identification, as a part of the i2b2 (Informatics for Integrating Biology to the Bedside) project, authors organized a Natural Language Processing (NLP) challenge on automatically removing private health information (PHI) from medical discharge records. This manuscript provides an overview of this de-identification challenge, describes the data and the annotation process, explains the evaluation metrics, discusses the nature of the systems that addressed the challenge, analyzes the results of received system runs, and identifies directions for future research. The de-indentification challenge data consisted of discharge summaries drawn from the Partners Healthcare system. Authors prepared this data for the challenge by replacing authentic PHI with synthesized surrogates. To focus the challenge on non-dictionary-based de-identification methods, the data was enriched with out-of-vocabulary PHI surrogates, i.e., made up names. The data also included some PHI surrogates that were ambiguous with medical non-PHI terms. A total of seven teams participated in the challenge. Each team submitted up to three system runs, for a total of sixteen submissions. The authors used precision, recall, and F-measure to evaluate the submitted system runs based on their token-level and instance-level performance on the ground truth. The systems with the best performance scored above 98% in F-measure for all categories of PHI. Most out-of-vocabulary PHI could be identified accurately. However, identifying ambiguous PHI proved challenging. The performance of systems on the test data set is encouraging. Future evaluations of these systems will involve larger data sets from more heterogeneous sources.
Figures






References
-
- Rollman B, Hanusa B, Gilbert T, Lowe H, Kapoor W, Schulberg H. The Electronic Medical Record Arch Intern Med 2001;161:89. - PubMed
-
- Cao H, Stetson P, Hripcsak G. Assessing Explicit Error Reporting in the Narrative Electronic Medical Record Using Keyword Searching J Biomed Inform 2004;36:99-105. - PubMed
-
- Chapman W, Bridewell W, Hanbury P, Cooper G, Buchanan B. A Simple Algorithm for Identifying Negated Findings and Diseases in Discharge Summaries J Biomed Inform 2001;34:301-310. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

