A single mutation in the active site swaps the substrate specificity of N-acetyl-L-ornithine transcarbamylase and N-succinyl-L-ornithine transcarbamylase
- PMID: 17600144
- PMCID: PMC2203365
- DOI: 10.1110/ps.072919907
A single mutation in the active site swaps the substrate specificity of N-acetyl-L-ornithine transcarbamylase and N-succinyl-L-ornithine transcarbamylase
Abstract
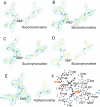
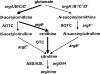
Transcarbamylases catalyze the transfer of the carbamyl group from carbamyl phosphate (CP) to an amino group of a second substrate such as aspartate, ornithine, or putrescine. Previously, structural determination of a transcarbamylase from Xanthomonas campestris led to the discovery of a novel N-acetylornithine transcarbamylase (AOTCase) that catalyzes the carbamylation of N-acetylornithine. Recently, a novel N-succinylornithine transcarbamylase (SOTCase) from Bacteroides fragilis was identified. Structural comparisons of AOTCase from X. campestris and SOTCase from B. fragilis revealed that residue Glu92 (X. campestris numbering) plays a critical role in distinguishing AOTCase from SOTCase. Enzymatic assays of E92P, E92S, E92V, and E92A mutants of AOTCase demonstrate that each of these mutations converts the AOTCase to an SOTCase. Similarly, the P90E mutation in B. fragilis SOTCase (equivalent to E92 in X. campestris AOTCase) converts the SOTCase to AOTCase. Hence, a single amino acid substitution is sufficient to swap the substrate specificities of AOTCase and SOTCase. X-ray crystal structures of these mutants in complexes with CP and N-acetyl-L-norvaline (an analog of N-acetyl-L-ornithine) or N-succinyl-L-norvaline (an analog of N-succinyl-L-ornithine) substantiate this conversion. In addition to Glu92 (X. campestris numbering), other residues such as Asn185 and Lys30 in AOTCase, which are involved in binding substrates through bridging water molecules, help to define the substrate specificity of AOTCase. These results provide the correct annotation (AOTCase or SOTCase) for a set of the transcarbamylase-like proteins that have been erroneously annotated as ornithine transcarbamylase (OTCase, EC 2.1.3.3).
Figures
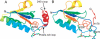

Similar articles
-
From Genome to Structure and Back Again: A Family Portrait of the Transcarbamylases.Int J Mol Sci. 2015 Aug 12;16(8):18836-64. doi: 10.3390/ijms160818836. Int J Mol Sci. 2015. PMID: 26274952 Free PMC article. Review.
-
Structure and catalytic mechanism of a novel N-succinyl-L-ornithine transcarbamylase in arginine biosynthesis of Bacteroides fragilis.J Biol Chem. 2006 Jul 21;281(29):20623-31. doi: 10.1074/jbc.M601229200. Epub 2006 May 16. J Biol Chem. 2006. PMID: 16704984
-
Structures of N-acetylornithine transcarbamoylase from Xanthomonas campestris complexed with substrates and substrate analogs imply mechanisms for substrate binding and catalysis.Proteins. 2006 Aug 1;64(2):532-42. doi: 10.1002/prot.21013. Proteins. 2006. PMID: 16741992
-
Crystal structure of N-acetylornithine transcarbamylase from Xanthomonas campestris: a novel enzyme in a new arginine biosynthetic pathway found in several eubacteria.J Biol Chem. 2005 Apr 15;280(15):14366-9. doi: 10.1074/jbc.C500005200. Epub 2005 Feb 24. J Biol Chem. 2005. PMID: 15731101
-
Sources and Fates of Carbamyl Phosphate: A Labile Energy-Rich Molecule with Multiple Facets.Biology (Basel). 2018 Jun 12;7(2):34. doi: 10.3390/biology7020034. Biology (Basel). 2018. PMID: 29895729 Free PMC article. Review.
Cited by
-
The ygeW encoded protein from Escherichia coli is a knotted ancestral catabolic transcarbamylase.Proteins. 2011 Jul;79(7):2327-34. doi: 10.1002/prot.23043. Epub 2011 May 9. Proteins. 2011. PMID: 21557323 Free PMC article.
-
Molecular cloning, characterization and positively selected sites of the glutathione S-transferase family from Locusta migratoria.PLoS One. 2014 Dec 8;9(12):e114776. doi: 10.1371/journal.pone.0114776. eCollection 2014. PLoS One. 2014. PMID: 25486043 Free PMC article.
-
Engineering the specificity of Streptococcus pyogenes sortase A by loop grafting.Proteins. 2020 Nov;88(11):1394-1400. doi: 10.1002/prot.25958. Epub 2020 Jun 21. Proteins. 2020. PMID: 32501594 Free PMC article.
-
Structural analysis and molecular substrate recognition properties of Arabidopsis thaliana ornithine transcarbamylase, the molecular target of phaseolotoxin produced by Pseudomonas syringae.Front Plant Sci. 2023 Dec 18;14:1297956. doi: 10.3389/fpls.2023.1297956. eCollection 2023. Front Plant Sci. 2023. PMID: 38179474 Free PMC article.
-
Characterization of the 5-hydroxymethylcytosine-specific DNA restriction endonucleases.Nucleic Acids Res. 2013 Apr;41(7):4198-206. doi: 10.1093/nar/gkt102. Epub 2013 Mar 12. Nucleic Acids Res. 2013. PMID: 23482393 Free PMC article.
References
-
- Allewell N.M., Shi, D., Morizono, H., and Tuchman, M. 1999. Molecular recognition by ornithine and aspartate transcarbamoylases. Acc. Chem. Res. 32: 885–894.
-
- Born T.L. and Blanchard, J.S. 1999. Structure/function studies on enzymes in the diaminopimelate pathway of bacterial cell wall biosynthesis. Curr. Opin. Chem. Biol. 3: 607–613. - PubMed
-
- Bradford M. 1976. Quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 1219–1223. - PubMed
-
- Brown C.J., Takayama, S., Campen, A.M., Vise, P., Marshall, T.W., Oldfield, C.J., Williams, C.J., and Dunker, A.K. 2002. Evolutionary rate heterogeneity in proteins with long disordered regions. J. Mol. Evol. 55: 104–110. - PubMed
-
- Brünger A.T. 1992. Free R value: A novel statistical quantity for assessing the accuracy of crystal structures. Nature 355: 472–475. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

