Persistence of attenuated HIV-1 rev alleles in an epidemiologically linked cohort of long-term survivors infected with nef-deleted virus
- PMID: 17601342
- PMCID: PMC1933581
- DOI: 10.1186/1742-4690-4-43
Persistence of attenuated HIV-1 rev alleles in an epidemiologically linked cohort of long-term survivors infected with nef-deleted virus
Abstract
Background: The Sydney blood bank cohort (SBBC) of long-term survivors consists of multiple individuals infected with nef-deleted, attenuated strains of human immunodeficiency virus type 1 (HIV-1). Although the cohort members have experienced differing clinical courses and now comprise slow progressors (SP) as well as long-term nonprogressors (LTNP), longitudinal analysis of nef/long-terminal repeat (LTR) sequences demonstrated convergent nef/LTR sequence evolution in SBBC SP and LTNP. Thus, the in vivo pathogenicity of attenuated HIV-1 strains harboured by SBBC members is dictated by factors other than nef/LTR. Therefore, to determine whether defects in other viral genes contribute to attenuation of these HIV-1 strains, we characterized dominant HIV-1 rev alleles that persisted in 4 SBBC subjects; C18, C64, C98 and D36.
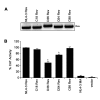
Results: The ability of Rev derived from D36 and C64 to bind the Rev responsive element (RRE) in RNA binding assays was reduced by approximately 90% compared to Rev derived from HIV-1NL4-3, C18 or C98. D36 Rev also had a 50-60% reduction in ability to express Rev-dependent reporter constructs in mammalian cells. In contrast, C64 Rev had only marginally decreased Rev function despite attenuated RRE binding. In D36 and C64, attenuated RRE binding was associated with rare amino acid changes at 3 highly conserved residues; Gln to Pro at position 74 immediately N-terminal to the Rev activation domain, and Val to Leu and Ser to Pro at positions 104 and 106 at the Rev C-terminus, respectively. In D36, reduced Rev function was mapped to an unusual 13 amino acid extension at the Rev C-terminus.
Conclusion: These findings provide new genetic and mechanistic insights important for Rev function, and suggest that Rev function, not Rev/RRE binding may be rate limiting for HIV-1 replication. In addition, attenuated rev alleles may contribute to viral attenuation and long-term survival of HIV-1 infection in a subset of SBBC members.
Figures


References
-
- Deacon NJ, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker DJ, McPhee DA, Greenway AL, Ellett A, Chatfield C, Lawson VA, Crowe S, Maertz S, Sonza S, Learmont J, Sullivan JS, Cunningham A, Dwyer D, Dowton D, Mills J. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. - DOI - PubMed
-
- Learmont JC, Geczy AF, Mills J, Ashton LJ, Raynes-Greenow CH, Garsia RJ, Dyer WB, McIntyre L, Oelrichs RB, Rhodes DI, Deacon NJ, Sullivan JS. Immunologic and virologic status after 14 to 18 years of infection with an attenuated strain of HIV-1. A report from the Sydney Blood Bank Cohort. N Engl J Med. 1999;340:1715–1722. doi: 10.1056/NEJM199906033402203. - DOI - PubMed
-
- Churchill MJ, Rhodes DI, Learmont JC, Sullivan JS, Wesselingh SL, Cooke IR, Deacon NJ, Gorry PR. Longitudinal analysis of human immunodeficiency virus type 1 nef/long terminal repeat sequences in a cohort of long-term survivors infected from a single source. J Virol. 2006;80:1047–1052. doi: 10.1128/JVI.80.2.1047-1052.2006. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

