Identification of a biosynthetic gene cluster and the six associated lipopeptides involved in swarming motility of Pseudomonas syringae pv. tomato DC3000
- PMID: 17601782
- PMCID: PMC1951903
- DOI: 10.1128/JB.00725-07
Identification of a biosynthetic gene cluster and the six associated lipopeptides involved in swarming motility of Pseudomonas syringae pv. tomato DC3000
Abstract
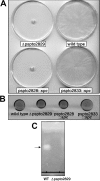
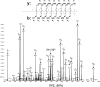
Pseudomonas species are known to be prolific producers of secondary metabolites that are synthesized wholly or in part by nonribosomal peptide synthetases. In an effort to identify additional nonribosomal peptides produced by these bacteria, a bioinformatics approach was used to "mine" the genome of Pseudomonas syringae pv. tomato DC3000 for the metabolic potential to biosynthesize previously unknown nonribosomal peptides. Herein we describe the identification of a nonribosomal peptide biosynthetic gene cluster that codes for proteins involved in the production of six structurally related linear lipopeptides. Structures for each of these lipopeptides were proposed based on amino acid analysis and mass spectrometry analyses. Mutations in this cluster resulted in the loss of swarming motility of P. syringae pv. tomato DC3000 on medium containing a low percentage of agar. This phenotype is consistent with the loss of the ability to produce a lipopeptide that functions as a biosurfactant. This work gives additional evidence that mining the genomes of microorganisms followed by metabolite and phenotypic analyses leads to the identification of previously unknown secondary metabolites.
Figures




References
-
- Balibar, C. J., F. H. Vaillancourt, and C. T. Walsh. 2005. Generation of D amino acid residues in assembly of arthrofactin by dual condensation/epimerization domains. Chem. Biol. 12:1189-1200. - PubMed
-
- Ballio, A., F. Bossa, D. Di Giorgio, A. Di Nola, C. Manetti, M. Paci, A. Scaloni, and A. L. Segre. 1995. Solution conformation of the Pseudomonas syringae pv. syringae phytotoxic lipodepsipeptide syringopeptin 25-A. Two-dimensional NMR, distance geometry and molecular dynamics. Eur. J. Biochem. 234:747-758. - PubMed
-
- Bergmann, S., J. Schumann, K. Scherlach, C. Lange, A. A. Brakhage, and C. Hertweck. 2007. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat. Chem. Biol. 3:213-217. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

