Reciprocal phosphorylation and glycosylation recognition motifs control NCAPP1 interaction with pumpkin phloem proteins and their cell-to-cell movement
- PMID: 17601822
- PMCID: PMC1955715
- DOI: 10.1105/tpc.107.052522
Reciprocal phosphorylation and glycosylation recognition motifs control NCAPP1 interaction with pumpkin phloem proteins and their cell-to-cell movement
Abstract
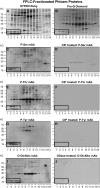
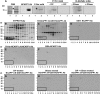
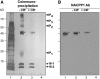
In plants, cell-to-cell trafficking of non-cell-autonomous proteins (NCAPs) involves protein-protein interactions, and a role for posttranslational modification has been implicated. In this study, proteins contained in pumpkin (Cucurbita maxima cv Big Max) phloem sap were used as a source of NCAPs to further explore the molecular basis for selective NCAP trafficking. Protein overlay assays and coimmunoprecipitation experiments established that phosphorylation and glycosylation, on both Nicotiana tabacum NON-CELL-AUTONOMOUS PATHWAY PROTEIN1 (Nt-NCAPP1) and the phloem NCAPs, are essential for their interaction. Detailed molecular analysis of a representative phloem NCAP, Cm-PP16-1, identified the specific residues on which glycosylation and phosphorylation must occur for effective binding to NCAPP1. Microinjection studies confirmed that posttranslational modification on these residues is essential for cell-to-cell movement of Cm-PP16-1. Lastly, a glutathione S-transferase (GST)-Cm-PP16-1 fusion protein system was employed to test whether the peptide region spanning these residues was required for cell-to-cell movement. These studies established that a 36-amino acid peptide was sufficient to impart cell-to-cell movement capacity to GST, a normally cell-autonomous protein. These findings are consistent with the hypothesis that a phosphorylation-glycosylation recognition motif functions to control the binding of a specific subset of phloem NCAPs to NCAPP1 and their subsequent transport through plasmodesmata.
Figures







Similar articles
-
Overexpression of Arabidopsis plasmodesmata germin-like proteins disrupts root growth and development.Plant Cell. 2012 Sep;24(9):3630-48. doi: 10.1105/tpc.112.101063. Epub 2012 Sep 7. Plant Cell. 2012. PMID: 22960910 Free PMC article.
-
Phloem long-distance delivery of FLOWERING LOCUS T (FT) to the apex.Plant J. 2013 Aug;75(3):456-68. doi: 10.1111/tpj.12213. Epub 2013 May 14. Plant J. 2013. PMID: 23607279
-
A polypyrimidine tract binding protein, pumpkin RBP50, forms the basis of a phloem-mobile ribonucleoprotein complex.Plant Cell. 2009 Jan;21(1):197-215. doi: 10.1105/tpc.108.061317. Epub 2009 Jan 2. Plant Cell. 2009. PMID: 19122103 Free PMC article.
-
Plasmodesmata - bridging the gap between neighboring plant cells.Trends Cell Biol. 2009 Oct;19(10):495-503. doi: 10.1016/j.tcb.2009.07.003. Epub 2009 Sep 10. Trends Cell Biol. 2009. PMID: 19748270 Review.
-
Mechanisms of phloem loading.Curr Opin Plant Biol. 2018 Jun;43:71-75. doi: 10.1016/j.pbi.2018.01.009. Epub 2018 Feb 12. Curr Opin Plant Biol. 2018. PMID: 29448176 Review.
Cited by
-
Trichoderma atroviride P1 Colonization of Tomato Plants Enhances Both Direct and Indirect Defense Barriers Against Insects.Front Physiol. 2019 Jul 5;10:813. doi: 10.3389/fphys.2019.00813. eCollection 2019. Front Physiol. 2019. PMID: 31333483 Free PMC article.
-
Macromolecular composition of phloem exudate from white lupin (Lupinus albus L.).BMC Plant Biol. 2011 Feb 22;11:36. doi: 10.1186/1471-2229-11-36. BMC Plant Biol. 2011. PMID: 21342527 Free PMC article.
-
O-GlcNAc protein modification in plants: Evolution and function.Biochim Biophys Acta. 2010 Feb;1800(2):49-56. doi: 10.1016/j.bbagen.2009.11.016. Epub 2009 Dec 2. Biochim Biophys Acta. 2010. PMID: 19961900 Free PMC article. Review.
-
A plasmodesmal glycosyltransferase-like protein.PLoS One. 2013;8(2):e58025. doi: 10.1371/journal.pone.0058025. Epub 2013 Feb 26. PLoS One. 2013. PMID: 23469135 Free PMC article.
-
Identification of lipids and lipid-binding proteins in phloem exudates from Arabidopsis thaliana.J Exp Bot. 2012 Jun;63(10):3603-16. doi: 10.1093/jxb/ers028. Epub 2012 Mar 21. J Exp Bot. 2012. PMID: 22442409 Free PMC article.
References
-
- Cao, Q., Kim, J.H., and Richter, J.D. (2006). CDK1 and calcineurin regulate Maskin association with eIF4E and translational control of cell cycle progression. Nat. Struct. Mol. Biol. 13 1128–1134. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

