LPA2 is required for efficient assembly of photosystem II in Arabidopsis thaliana
- PMID: 17601825
- PMCID: PMC1955735
- DOI: 10.1105/tpc.107.050526
LPA2 is required for efficient assembly of photosystem II in Arabidopsis thaliana
Retraction in
-
RETRACTION.Plant Cell. 2016 Dec;28(12):3061. doi: 10.1105/tpc.16.00881. Epub 2016 Nov 28. Plant Cell. 2016. PMID: 27895223 Free PMC article. No abstract available.
Abstract
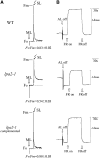
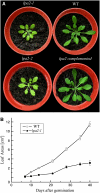
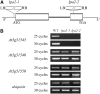
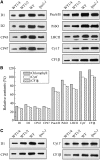
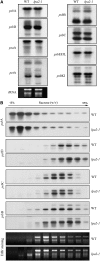
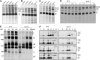
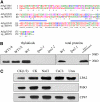
To elucidate the molecular mechanism of photosystem II (PSII) assembly, we characterized the low psii accumulation2 (lpa2) mutant of Arabidopsis thaliana, which is defective in the accumulation of PSII supercomplexes. The levels and processing patterns of the RNAs encoding the PSII subunits are unaltered in the mutant. In vivo protein-labeling experiments showed that the synthesis of CP43 (for chlorophyll a binding protein) was greatly reduced, but CP47, D1, and D2 were synthesized at normal rates in the lpa2-1 mutant. The newly synthesized CP43 was rapidly degraded in lpa2-1, and the turnover rates of D1 and D2 were higher in lpa2-1 than in wild-type plants. The newly synthesized PSII proteins were assembled into PSII complexes, but the assembly of PSII was less efficient in the mutant than in wild-type plants. LPA2 encodes an intrinsic thylakoid membrane protein, which is not an integral subunit of PSII. Yeast two-hybrid assays indicated that LPA2 interacts with the PSII core protein CP43 but not with the PSII reaction center proteins D1 and D2. Moreover, direct interactions of LPA2 with Albino3 (Alb3), which is involved in thylakoid membrane biogenesis and cell division, were also detected. Thus, the results suggest that LPA2, which appears to form a complex with Alb3, is involved in assisting CP43 assembly within PSII.
Figures



Similar articles
-
The Arabidopsis thylakoid protein PAM68 is required for efficient D1 biogenesis and photosystem II assembly.Plant Cell. 2010 Oct;22(10):3439-60. doi: 10.1105/tpc.110.077453. Epub 2010 Oct 5. Plant Cell. 2010. PMID: 20923938 Free PMC article.
-
Cooperation of LPA3 and LPA2 is essential for photosystem II assembly in Arabidopsis.Plant Physiol. 2010 Sep;154(1):109-20. doi: 10.1104/pp.110.159558. Epub 2010 Jul 6. Plant Physiol. 2010. Retraction in: Plant Physiol. 2017 Feb;173(2):1526. doi: 10.1104/pp.16.01908. PMID: 20605914 Free PMC article. Retracted.
-
The Arabidopsis Tellurite resistance C protein together with ALB3 is involved in photosystem II protein synthesis.Plant J. 2014 Apr;78(2):344-56. doi: 10.1111/tpj.12474. Epub 2014 Mar 26. Plant J. 2014. PMID: 24612058
-
Processing of D1 Protein: A Mysterious Process Carried Out in Thylakoid Lumen.Int J Mol Sci. 2022 Feb 25;23(5):2520. doi: 10.3390/ijms23052520. Int J Mol Sci. 2022. PMID: 35269663 Free PMC article. Review.
-
The biogenesis and maintenance of PSII: Recent advances and current challenges.Plant Cell. 2024 Oct 3;36(10):3997-4013. doi: 10.1093/plcell/koae082. Plant Cell. 2024. PMID: 38484127 Free PMC article. Review.
Cited by
-
Evolution of plant senescence.BMC Evol Biol. 2009 Jul 14;9:163. doi: 10.1186/1471-2148-9-163. BMC Evol Biol. 2009. PMID: 19602260 Free PMC article.
-
HYPERSENSITIVE TO HIGH LIGHT1 interacts with LOW QUANTUM YIELD OF PHOTOSYSTEM II1 and functions in protection of photosystem II from photodamage in Arabidopsis.Plant Cell. 2014 Mar;26(3):1213-29. doi: 10.1105/tpc.113.122424. Epub 2014 Mar 14. Plant Cell. 2014. PMID: 24632535 Free PMC article.
-
Cost and benefit of the repair of photodamaged photosystem II in spinach leaves: roles of acclimation to growth light.Photosynth Res. 2012 Sep;113(1-3):165-80. doi: 10.1007/s11120-012-9767-0. Epub 2012 Jul 15. Photosynth Res. 2012. PMID: 22797856
-
The Arabidopsis thylakoid protein PAM68 is required for efficient D1 biogenesis and photosystem II assembly.Plant Cell. 2010 Oct;22(10):3439-60. doi: 10.1105/tpc.110.077453. Epub 2010 Oct 5. Plant Cell. 2010. PMID: 20923938 Free PMC article.
-
A comparative study of wavelength-dependent photoinactivation in photosystem II of drought-tolerant photosynthetic organisms in Antarctica and the potential risks of photoinhibition in the habitat.Ann Bot. 2018 Dec 31;122(7):1263-1278. doi: 10.1093/aob/mcy139. Ann Bot. 2018. PMID: 30052754 Free PMC article.
References
-
- Adir, N., Schochat, S., and Ohad, I. (1990). Light-dependent D1 protein synthesis and translocation is regulated by reaction centre II. Reaction centre II serves as an acceptor for the D1 precursor. J. Biol. Chem. 265 12563–12568. - PubMed
-
- Barkan, A., and Goldschmidt-Clermont, M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82 559–572. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

