Expression profiling of Dexamethasone-treated primary chondrocytes identifies targets of glucocorticoid signalling in endochondral bone development
- PMID: 17603917
- PMCID: PMC1929075
- DOI: 10.1186/1471-2164-8-205
Expression profiling of Dexamethasone-treated primary chondrocytes identifies targets of glucocorticoid signalling in endochondral bone development
Abstract
Background: Glucocorticoids (GCs) are widely used anti-inflammatory drugs. While useful in clinical practice, patients taking GCs often suffer from skeletal side effects including growth retardation in children and adolescents, and decreased bone quality in adults. On a physiological level, GCs have been implicated in the regulation of chondrogenesis and osteoblast differentiation, as well as maintaining homeostasis in cartilage and bone. We identified the glucocorticoid receptor (GR) as a potential regulator of chondrocyte hypertrophy in a microarray screen of primary limb bud mesenchyme micromass cultures. Some targets of GC regulation in chondrogenesis are known, but the global effects of pharmacological GC doses on chondrocyte gene expression have not been comprehensively evaluated.
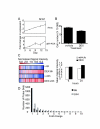
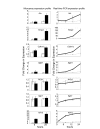
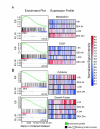
Results: This study systematically identifies a spectrum of GC target genes in embryonic growth plate chondrocytes treated with a synthetic GR agonist, dexamethasone (DEX), at 6 and 24 hrs. Conventional analysis of this data set and gene set enrichment analysis (GSEA) was performed. Transcripts associated with metabolism were enriched in the DEX condition along with extracellular matrix genes. In contrast, a subset of growth factors and cytokines were negatively correlated with DEX treatment. Comparing DEX-induced gene expression data to developmental changes in gene expression in micromass cultures revealed an additional layer of complexity in which DEX maintains the expression of certain chondrocyte marker genes while inhibiting factors that promote vascularization and ultimately ossification of the cartilaginous template.
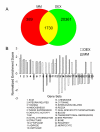
Conclusion: Together, these results provide insight into the mechanisms and major molecular classes functioning downstream of DEX in primary chondrocytes. In addition, comparison of our data with microarray studies of DEX treatment in other cell types demonstrated that the majority of DEX effects are tissue-specific. This study provides novel insights into the effects of pharmacological GC on chondrocyte gene transcription and establishes the foundation for subsequent functional studies.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

