Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs
- PMID: 17604720
- PMCID: PMC2084369
- DOI: 10.1016/j.cell.2007.05.022
Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs
Abstract
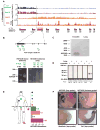
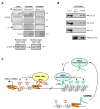
Noncoding RNAs (ncRNA) participate in epigenetic regulation but are poorly understood. Here we characterize the transcriptional landscape of the four human HOX loci at five base pair resolution in 11 anatomic sites and identify 231 HOX ncRNAs that extend known transcribed regions by more than 30 kilobases. HOX ncRNAs are spatially expressed along developmental axes and possess unique sequence motifs, and their expression demarcates broad chromosomal domains of differential histone methylation and RNA polymerase accessibility. We identified a 2.2 kilobase ncRNA residing in the HOXC locus, termed HOTAIR, which represses transcription in trans across 40 kilobases of the HOXD locus. HOTAIR interacts with Polycomb Repressive Complex 2 (PRC2) and is required for PRC2 occupancy and histone H3 lysine-27 trimethylation of HOXD locus. Thus, transcription of ncRNA may demarcate chromosomal domains of gene silencing at a distance; these results have broad implications for gene regulation in development and disease states.
Figures





Comment in
-
HOTAIR lifts noncoding RNAs to new levels.Cell. 2007 Jun 29;129(7):1257-9. doi: 10.1016/j.cell.2007.06.014. Cell. 2007. PMID: 17604716 Review.
References
-
- Albrecht UHJA, Lin H. Visualization of gene expression patterns by in situ hybridization. Boca Raton, FL: CRC Press; 1997.
-
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert J, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, et al. Cell. Vol. 120. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse; pp. 169–181. - PubMed
-
- Bernstein E, Allis CD. RNA meets chromatin. Genes Dev. 2005;19:1635–1655. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

