A mammalian microRNA expression atlas based on small RNA library sequencing
- PMID: 17604727
- PMCID: PMC2681231
- DOI: 10.1016/j.cell.2007.04.040
A mammalian microRNA expression atlas based on small RNA library sequencing
Abstract
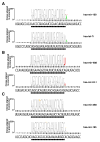
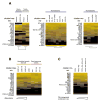
MicroRNAs (miRNAs) are small noncoding regulatory RNAs that reduce stability and/or translation of fully or partially sequence-complementary target mRNAs. In order to identify miRNAs and to assess their expression patterns, we sequenced over 250 small RNA libraries from 26 different organ systems and cell types of human and rodents that were enriched in neuronal as well as normal and malignant hematopoietic cells and tissues. We present expression profiles derived from clone count data and provide computational tools for their analysis. Unexpectedly, a relatively small set of miRNAs, many of which are ubiquitously expressed, account for most of the differences in miRNA profiles between cell lineages and tissues. This broad survey also provides detailed and accurate information about mature sequences, precursors, genome locations, maturation processes, inferred transcriptional units, and conservation patterns. We also propose a subclassification scheme for miRNAs for assisting future experimental and computational functional analyses.
Figures



Comment in
-
Mustering the micromanagers.Nat Biotechnol. 2007 Sep;25(9):996-7. doi: 10.1038/nbt0907-996. Nat Biotechnol. 2007. PMID: 17846629 No abstract available.
References
-
- Aravin A, Tuschl T. Identification and characterization of small RNAs involved in RNA silencing. FEBS Lett. 2005;579:5830–5840. - PubMed
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

