Widespread, exceptionally high levels of histone H3 lysine 4 trimethylation largely mediate "privileged" gene expression
- PMID: 17605300
- PMCID: PMC6032458
- DOI: 10.3727/000000006780666966
Widespread, exceptionally high levels of histone H3 lysine 4 trimethylation largely mediate "privileged" gene expression
Abstract
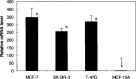
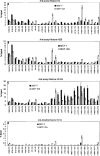
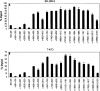
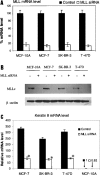
We examined the molecular determinants for sustained high-level expression of "privileged" genes, defined as the 0.03% most highly expressed genes within any specific cell. We identified histone modifications by chromatin immunoprecipitation analyses on Keratin 8, the most highly expressed gene in the human breast cancer cell line, MCF-7, based on serial analysis of gene expression. Quantitative comparisons to the "normal" counterpart cell line, MCF-10A, expressing 350-fold lower levels of Keratin 8 and other breast cancer cell lines expressing higher levels were performed using real-time PCR. Extraordinarily high levels of trimethyl histone H3 lysine 4 (H3K4) were found primarily in the first intron of the Keratin 8 gene stretching from 400 to 2000 bp downstream from the promoter in all breast cancer cells lines but not in MCF-10A cells. The highest levels of histone H3K4 trimethylation in MCF-7 cells ranged from 70% to 80% over input within 1200 bp of this region. Knockdown of mixed-lineage leukemia (MLL), the specific methyltransferase for histone H3K4, with MLL-specific siRNA decreased histone H3K4 trimethylation on the Keratin 8 gene and decreased Keratin 8 mRNA levels. Histone H3K4 trimethylation mediates approximately 86% of the elevated, sustained expression of the Keratin 8 gene in MCF-7 cells.
Figures


Similar articles
-
Differentially expressed genes are marked by histone 3 lysine 9 trimethylation in human cancer cells.Oncogene. 2008 Apr 10;27(17):2412-21. doi: 10.1038/sj.onc.1210895. Epub 2007 Oct 29. Oncogene. 2008. PMID: 17968314
-
Dynamic association of MLL1, H3K4 trimethylation with chromatin and Hox gene expression during the cell cycle.FEBS J. 2009 Mar;276(6):1629-40. doi: 10.1111/j.1742-4658.2009.06895.x. Epub 2009 Feb 7. FEBS J. 2009. PMID: 19220463
-
Arabidopsis COMPASS-like complexes mediate histone H3 lysine-4 trimethylation to control floral transition and plant development.PLoS Genet. 2011 Mar;7(3):e1001330. doi: 10.1371/journal.pgen.1001330. Epub 2011 Mar 10. PLoS Genet. 2011. PMID: 21423667 Free PMC article.
-
Histone H3K4 trimethylation: dynamic interplay with pre-mRNA splicing.Biochem Cell Biol. 2016 Feb;94(1):1-11. doi: 10.1139/bcb-2015-0065. Epub 2015 Jul 15. Biochem Cell Biol. 2016. PMID: 26352678 Review.
-
The cancer driver genes IDH1/2, JARID1C/ KDM5C, and UTX/ KDM6A: crosstalk between histone demethylation and hypoxic reprogramming in cancer metabolism.Exp Mol Med. 2019 Jun 20;51(6):1-17. doi: 10.1038/s12276-019-0230-6. Exp Mol Med. 2019. PMID: 31221981 Free PMC article. Review.
Cited by
-
Renal ischemia-induced cholesterol loading: transcription factor recruitment and chromatin remodeling along the HMG CoA reductase gene.Am J Pathol. 2009 Jan;174(1):54-62. doi: 10.2353/ajpath.2009.080602. Epub 2008 Dec 18. Am J Pathol. 2009. PMID: 19095962 Free PMC article.
-
Endotoxin mediates recruitment of RNA polymerase II to target genes in acute renal failure.J Am Soc Nephrol. 2008 Jul;19(7):1321-30. doi: 10.1681/ASN.2007121368. Epub 2008 Apr 16. J Am Soc Nephrol. 2008. PMID: 18417719 Free PMC article.
-
Optimization of Non-Viral Gene Therapeutics Using Bilamellar Invaginated Vesicles.J Genet Syndr Gene Ther. 2011 Dec 17;(S5):002. doi: 10.4172/2157-7412.s5-002. J Genet Syndr Gene Ther. 2011. PMID: 23087840 Free PMC article.
-
Chromatin dynamics: H3K4 methylation and H3 variant replacement during development and in cancer.Cell Mol Life Sci. 2014 Sep;71(18):3439-63. doi: 10.1007/s00018-014-1605-4. Epub 2014 Mar 28. Cell Mol Life Sci. 2014. PMID: 24676717 Free PMC article. Review.
References
-
- Bernstein B. E.; Kamal M.; Lindblad-Toh K.; Bekiranov S.; Bailey D. K.; Huebert D. J.; McMahon S.; Karlsson E. K.; Kulbokas E. J. III; Gingeras T. R.; Schreiber S. L.; Lander E. S. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120:169–181; 2005. - PubMed
-
- Brotherick I.; Robson C. N.; Browell D. A.; Shenfine J.; White M. D.; Cunliffe W. J.; Shenton B. K.; Egan M.; Webb L. A.; Lunt L. G.; Young J. R.; Higgs M. J. Cytokeratin expression in breast cancer: Phenotypic changes associated with disease progression. Cytometry 32:301–308; 1998. - PubMed
-
- Cianga C.; Cianga P.; Cozma L.; Diaconu C.; Carasevici E. Detection of lymph nodes micrometastases in breast carcinoma using immunohistochemistry for cytokeratin 8. Rev. Med. Chir. Soc. Med. Nat. Iasi 106:720–724; 2002. - PubMed
-
- Godfroid E.; Geuskens M.; Dupressoir T.; Parent I.; Szpirer C. Cytokeratins are exposed on the outer surface of established human mammary carcinoma cells. J. Cell Sci. 99:595–607; 1991. - PubMed
-
- Gonias S. L.; Hembrough T. A.; Sankovic M. Cytokeratin 8 functions as a major plasminogen receptor in select epithelial and carcinoma cells. Front. Biosci. 6:D1403–D1411; 2001. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
