Small but versatile: the extraordinary functional and structural diversity of the beta-grasp fold
- PMID: 17605815
- PMCID: PMC1949818
- DOI: 10.1186/1745-6150-2-18
Small but versatile: the extraordinary functional and structural diversity of the beta-grasp fold
Abstract
Background: The beta-grasp fold (beta-GF), prototyped by ubiquitin (UB), has been recruited for a strikingly diverse range of biochemical functions. These functions include providing a scaffold for different enzymatic active sites (e.g. NUDIX phosphohydrolases) and iron-sulfur clusters, RNA-soluble-ligand and co-factor-binding, sulfur transfer, adaptor functions in signaling, assembly of macromolecular complexes and post-translational protein modification. To understand the basis for the functional versatility of this small fold we undertook a comprehensive sequence-structure analysis of the fold and developed a natural classification for its members.
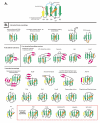
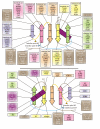
Results: As a result we were able to define the core distinguishing features of the fold and numerous elaborations, including several previously unrecognized variants. Systematic analysis of all known interactions of the fold showed that its manifold functional abilities arise primarily from the prominent beta-sheet, which provides an exposed surface for diverse interactions or additionally, by forming open barrel-like structures. We show that in the beta-GF both enzymatic activities and the binding of diverse co-factors (e.g. molybdopterin) have independently evolved on at least three occasions each, and iron-sulfur-cluster-binding on at least two independent occasions. Our analysis identified multiple previously unknown large monophyletic assemblages within the beta-GF, including one which unifies versions found in the fasciclin-1 superfamily, the ribosomal protein L25, the phosphoribosyl AMP cyclohydrolase (HisI) and glutamine synthetase. We also uncovered several new groups of beta-GF domains including a domain found in bacterial flagellar and fimbrial assembly components, and 5 new UB-like domains in the eukaryotes.
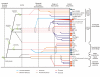
Conclusion: Evolutionary reconstruction indicates that the beta-GF had differentiated into at least 7 distinct lineages by the time of the last universal common ancestor of all extant organisms, encompassing much of the structural diversity observed in extant versions of the fold. The earliest beta-GF members were probably involved in RNA metabolism and subsequently radiated into various functional niches. Most of the structural diversification occurred in the prokaryotes, whereas the eukaryotic phase was mainly marked by a specific expansion of the ubiquitin-like beta-GF members. The eukaryotic UB superfamily diversified into at least 67 distinct families, of which at least 19-20 families were already present in the eukaryotic common ancestor, including several protein and one lipid conjugated forms. Another key aspect of the eukaryotic phase of evolution of the beta-GF was the dramatic increase in domain architectural complexity of proteins related to the expansion of UB-like domains in numerous adaptor roles.
Figures



References
-
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

