A high-accuracy consensus map of yeast protein complexes reveals modular nature of gene essentiality
- PMID: 17605818
- PMCID: PMC1940025
- DOI: 10.1186/1471-2105-8-236
A high-accuracy consensus map of yeast protein complexes reveals modular nature of gene essentiality
Abstract
Background: Identifying all protein complexes in an organism is a major goal of systems biology. In the past 18 months, the results of two genome-scale tandem affinity purification-mass spectrometry (TAP-MS) assays in yeast have been published, along with corresponding complex maps. For most complexes, the published data sets were surprisingly uncorrelated. It is therefore useful to consider the raw data from each study and generate an accurate complex map from a high-confidence data set that integrates the results of these and earlier assays.
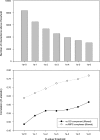
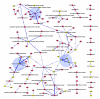
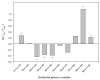
Results: Using an unsupervised probabilistic scoring scheme, we assigned a confidence score to each interaction in the matrix-model interpretation of the large-scale yeast mass-spectrometry data sets. The scoring metric proved more accurate than the filtering schemes used in the original data sets. We then took a high-confidence subset of these interactions and derived a set of complexes using MCL. The complexes show high correlation with existing annotations. Hierarchical organization of some protein complexes is evident from inter-complex interactions.
Conclusion: We demonstrate that our scoring method can generate an integrated high-confidence subset of observed matrix-model interactions, which we subsequently used to derive an accurate map of yeast complexes. Our results indicate that essentiality is a product of the protein complex rather than the individual protein, and that we have achieved near saturation of the yeast high-abundance, rich-media-expressed "complex-ome."
Figures


References
-
- Reguly T, Breitkreutz A, Boucher L, Breitkreutz BJ, Hon GC, Myers CL, Parsons A, Friesen H, Oughtred R, Tong A, Stark C, Ho Y, Botstein D, Andrews B, Boone C, Troyanskya OG, Ideker T, Dolinski K, Batada NN, Tyers M. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J Biol. 2006;5:11. doi: 10.1186/jbiol36. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

