Ets factors and a newly identified polymorphism regulate Fli1 promoter activity in lymphocytes
- PMID: 17606295
- PMCID: PMC2045641
- DOI: 10.1016/j.molimm.2007.05.018
Ets factors and a newly identified polymorphism regulate Fli1 promoter activity in lymphocytes
Abstract
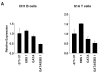
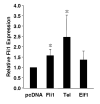
Fli1 is an Ets family member that is essential for embryonic development. Increasing evidence suggests modulating Fli1 gene expression impacts lymphocyte development/function and is an important mediator in the autoimmune disease lupus. Fli1 is over-expressed in splenic lymphocytes in lupus prone mouse strains and in PBMCs of lupus patients. Presently, it is unknown how Fli1 gene expression is controlled in lymphocytes or how it becomes over-expressed in lupus. Therefore, we examined Fli1 regulation in a murine B cell line and T cell line and identified several cis-regulatory elements within a 230 bp region that contribute to Fli1 promoter activity. Ets factors Elf1, Tel and Fli1 bind in vitro to this region and increase endogenous Fli1 expression when over-expressed in a T cell line. In addition, we determined that a microsatellite located adjacent to the region containing these cis-regulatory elements is polymorphic in three lupus prone mouse strains and that the length of the microsatellite is inversely correlated with promoter activity in a T cell line. These results suggest that several Ets factors, including Fli1 itself, are involved in the transcriptional regulation of Fli1 in lymphocytes. Furthermore, the presence of a polymorphic microsatellite in the Fli1 promoter may contribute to increased Fli1 expression in T cells during lupus disease progression.
Figures











Similar articles
-
A GA microsatellite in the Fli1 promoter modulates gene expression and is associated with systemic lupus erythematosus patients without nephritis.Arthritis Res Ther. 2010;12(6):R212. doi: 10.1186/ar3189. Epub 2010 Nov 18. Arthritis Res Ther. 2010. PMID: 21087477 Free PMC article.
-
The mouse and human Fli1 genes are similarly regulated by Ets factors in T cells.Genes Immun. 2010 Mar;11(2):161-72. doi: 10.1038/gene.2009.73. Epub 2009 Oct 15. Genes Immun. 2010. PMID: 19829305 Free PMC article.
-
FLI1 and EWS-FLI1 function as ternary complex factors and ELK1 and SAP1a function as ternary and quaternary complex factors on the Egr1 promoter serum response elements.Oncogene. 1997 Jan 16;14(2):213-21. doi: 10.1038/sj.onc.1200839. Oncogene. 1997. PMID: 9010223
-
Fli1, Elf1, and Ets1 regulate the proximal promoter of the LMO2 gene in endothelial cells.Blood. 2005 Oct 15;106(8):2680-7. doi: 10.1182/blood-2004-12-4755. Epub 2005 Jun 30. Blood. 2005. PMID: 15994290
-
Regulation of Fli1 gene expression and lupus.Autoimmun Rev. 2006 Jul;5(6):377-82. doi: 10.1016/j.autrev.2005.10.005. Epub 2005 Dec 13. Autoimmun Rev. 2006. PMID: 16890890 Review.
Cited by
-
Fli-1 Activation through Targeted Promoter Activity Regulation Using a Novel 3', 5'-diprenylated Chalcone Inhibits Growth and Metastasis of Prostate Cancer Cells.Int J Mol Sci. 2020 Mar 23;21(6):2216. doi: 10.3390/ijms21062216. Int J Mol Sci. 2020. PMID: 32210104 Free PMC article.
-
Fine mapping of Leishmania major susceptibility Locus lmr2 and evidence of a role for Fli1 in disease and wound healing.Infect Immun. 2010 Jun;78(6):2734-44. doi: 10.1128/IAI.00126-10. Epub 2010 Apr 5. Infect Immun. 2010. PMID: 20368343 Free PMC article.
-
A GA microsatellite in the Fli1 promoter modulates gene expression and is associated with systemic lupus erythematosus patients without nephritis.Arthritis Res Ther. 2010;12(6):R212. doi: 10.1186/ar3189. Epub 2010 Nov 18. Arthritis Res Ther. 2010. PMID: 21087477 Free PMC article.
-
The mouse and human Fli1 genes are similarly regulated by Ets factors in T cells.Genes Immun. 2010 Mar;11(2):161-72. doi: 10.1038/gene.2009.73. Epub 2009 Oct 15. Genes Immun. 2010. PMID: 19829305 Free PMC article.
-
Understanding the role of ETS-mediated gene regulation in complex biological processes.Adv Cancer Res. 2013;119:1-61. doi: 10.1016/B978-0-12-407190-2.00001-0. Adv Cancer Res. 2013. PMID: 23870508 Free PMC article. Review.
References
-
- Anderson MK, Hernandez-Hoyos G, Diamond RA, Rothenberg EV. Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development. 1999;126:3131–48. - PubMed
-
- Barat C, Barbeau B, Delattre O, Rassart E. Presence of new alternative exons in human and mouse Fli-1 genes. Biochim Biophys Acta. 2000;1517:164–70. - PubMed
-
- Barbeau B, Barat C, Bergeron D, Rassart E. The GATA-1 and Spi-1 transcriptional factors bind to a GATA/EBS dual element in the Fli-1 exon 1. Oncogene. 1999;18:5535–45. - PubMed
-
- Barbeau B, Bergeron D, Beaulieu M, Nadjem Z, Rassart E. Characterization of the human and mouse Fli-1 promoter regions. Biochim Biophys Acta. 1996;1307:220–32. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

