A nonsense mutation in agrA accounts for the defect in agr expression and the avirulence of Staphylococcus aureus 8325-4 traP::kan
- PMID: 17606604
- PMCID: PMC1951176
- DOI: 10.1128/IAI.00679-07
A nonsense mutation in agrA accounts for the defect in agr expression and the avirulence of Staphylococcus aureus 8325-4 traP::kan
Abstract
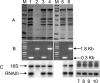
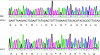
TraP is a triply phosphorylated staphylococcal protein that has been hypothesized to be the mediator of a second Staphylococcus aureus quorum-sensing system, "SQS1," that controls expression of the agr system and therefore is essential for the organism's virulence. This hypothesis was based on the loss of agr expression and virulence by a traP mutant of strain 8325-4 and was supported by full complementation of both phenotypic defects by the cloned traP gene in strain NB8 (Y. Gov, I. Borovok, M. Korem, V. K. Singh, R. K. Jayaswal, B. J. Wilkinson, S. M. Rich, and N. Balaban, J. Biol. Chem. 279:14665-14672, 2004), in which the wild-type traP gene was expressed in trans in the 8325-4 traP mutant. We initiated a study of the mechanism by which TraP activates agr and found that the traP mutant strain used for this and other recently published studies has a second mutation, an adventitious stop codon in the middle of agrA, the agr response regulator. The traP mutation, once separated from the agrA defect by outcrossing, had no effect on agr expression or virulence, indicating that the agrA defect accounts fully for the lack of agr expression and for the loss of virulence attributed to the traP mutation. In addition, DNA sequencing showed that the agrA gene in strain NB8 (Gov et al., J. Biol. Chem., 2004), in contrast to that in the agr-defective 8325-4 traP mutant strain, had the wild-type sequence; further, the traP mutation in that strain, when outcrossed, also had no effect on agr expression.
Figures




References
-
- Arvidson, S., and K. Tegmark. 2001. Regulation of virulence determinants in Staphylococcus aureus. Int. J. Med. Microbiol. 291:159-170. - PubMed
-
- Balaban, N., T. Goldkorn, Y. Gov, M. Hirshberg, N. Koyfman, H. R. Matthews, R. T. Nhan, B. Singh, and O. Uziel. 2001. Regulation of Staphylococcus aureus pathogenesis via target of RNAIII-activating protein (TRAP). J. Biol. Chem. 276:2658-2667. - PubMed
-
- Cheung, A. L., and G. Zhang. 2002. Global regulation of virulence determinants in Staphylococcus aureus by the SarA protein family. Front. Biosci. 7:1825-1842. - PubMed
-
- Gill, S. R., D. E. Fouts, G. L. Archer, E. F. Mongodin, R. T. Deboy, J. Ravel, I. T. Paulsen, J. F. Kolonay, L. Brinkac, M. Beanan, R. J. Dodson, S. C. Daugherty, R. Madupu, S. V. Angiuoli, A. S. Durkin, D. H. Haft, J. Vamathevan, H. Khouri, T. Utterback, C. Lee, G. Dimitrov, L. Jiang, H. Qin, J. Weidman, K. Tran, K. Kang, I. R. Hance, K. E. Nelson, and C. M. Fraser. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426-2438. - PMC - PubMed
-
- Gov, Y., I. Borovok, M. Korem, V. K. Singh, R. K. Jayaswal, B. J. Wilkinson, S. M. Rich, and N. Balaban. 2004. Quorum sensing in staphylococci is regulated via phosphorylation of three conserved histidine residues. J. Biol. Chem. 279:14665-14672. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

