The 2'-O-ribose methyltransferase for cap 1 of spliced leader RNA and U1 small nuclear RNA in Trypanosoma brucei
- PMID: 17606627
- PMCID: PMC1952150
- DOI: 10.1128/MCB.00647-07
The 2'-O-ribose methyltransferase for cap 1 of spliced leader RNA and U1 small nuclear RNA in Trypanosoma brucei
Abstract
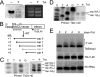
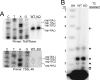
mRNA cap 1 2'-O-ribose methylation is a widespread modification that is implicated in processing, trafficking, and translational control in eukaryotic systems. The eukaryotic enzyme has yet to be identified. In kinetoplastid flagellates trans-splicing of spliced leader (SL) to polycistronic precursors conveys a hypermethylated cap 4, including a cap 0 m7G and seven additional methylations on the first 4 nucleotides, to all nuclear mRNAs. We report the first eukaryotic cap 1 2'-O-ribose methyltransferase, TbMTr1, a member of a conserved family of viral and eukaryotic enzymes. Recombinant TbMTr1 methylates the ribose of the first nucleotide of an m7G-capped substrate. Knockdowns and null mutants of TbMTr1 in Trypanosoma brucei grow normally, with loss of 2'-O-ribose methylation at cap 1 on substrate SL RNA and U1 small nuclear RNA. TbMTr1-null cells have an accumulation of cap 0 substrate without further methylation, while spliced mRNA is modified efficiently at position 4 in the absence of 2'-O-ribose methylation at position 1; downstream cap 4 methylations are independent of cap 1. Based on TbMTr1-green fluorescent protein localization, 2'-O-ribose methylation at position 1 occurs in the nucleus. Accumulation of 3'-extended SL RNA substrate indicates a delay in processing and suggests a synergistic role for cap 1 in maturation.
Figures




Similar articles
-
The TbMTr1 spliced leader RNA cap 1 2'-O-ribose methyltransferase from Trypanosoma brucei acts with substrate specificity.J Biol Chem. 2008 Feb 8;283(6):3161-3172. doi: 10.1074/jbc.M707367200. Epub 2007 Nov 29. J Biol Chem. 2008. PMID: 18048356
-
Trypanosoma brucei spliced leader RNA maturation by the cap 1 2'-O-ribose methyltransferase and SLA1 H/ACA snoRNA pseudouridine synthase complex.Mol Cell Biol. 2009 Mar;29(5):1202-11. doi: 10.1128/MCB.01496-08. Epub 2008 Dec 22. Mol Cell Biol. 2009. PMID: 19103757 Free PMC article.
-
Complete cap 4 formation is not required for viability in Trypanosoma brucei.Eukaryot Cell. 2006 Jun;5(6):905-15. doi: 10.1128/EC.00080-06. Eukaryot Cell. 2006. PMID: 16757738 Free PMC article.
-
Unconventional rules of small nuclear RNA transcription and cap modification in trypanosomatids.Gene Expr. 2002;10(1-2):3-16. Gene Expr. 2002. PMID: 11868986 Free PMC article. Review.
-
mRNA splicing in trypanosomes.Int J Med Microbiol. 2012 Oct;302(4-5):221-4. doi: 10.1016/j.ijmm.2012.07.004. Epub 2012 Sep 7. Int J Med Microbiol. 2012. PMID: 22964417 Review.
Cited by
-
Nuclear mRNA maturation and mRNA export control: from trypanosomes to opisthokonts.Parasitology. 2021 Sep;148(10):1196-1218. doi: 10.1017/S0031182021000068. Epub 2021 Jan 19. Parasitology. 2021. PMID: 33461637 Free PMC article. Review.
-
Characterization of hMTr1, a human Cap1 2'-O-ribose methyltransferase.J Biol Chem. 2010 Oct 22;285(43):33037-33044. doi: 10.1074/jbc.M110.155283. Epub 2010 Aug 16. J Biol Chem. 2010. PMID: 20713356 Free PMC article.
-
Structural analysis of human 2'-O-ribose methyltransferases involved in mRNA cap structure formation.Nat Commun. 2014;5:3004. doi: 10.1038/ncomms4004. Nat Commun. 2014. PMID: 24402442 Free PMC article.
-
Kinetoplastid genomics: the thin end of the wedge.Infect Genet Evol. 2008 Dec;8(6):901-6. doi: 10.1016/j.meegid.2008.07.001. Epub 2008 Jul 15. Infect Genet Evol. 2008. PMID: 18675383 Free PMC article.
-
Trypanosome mRNA recapping is triggered by hypermethylation originating from cap 4.Nucleic Acids Res. 2024 Sep 23;52(17):10645-10653. doi: 10.1093/nar/gkae614. Nucleic Acids Res. 2024. PMID: 39011881 Free PMC article.
References
-
- Alexandrov, A., I. Chernyakov, W. Gu, S. L. Hiley, T. R. Hughes, E. J. Grayhack, and E. M. Phizicky. 2006. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell 21:87-96. - PubMed
-
- Arhin, G. K., E. Ullu, and C. Tschudi. 2006. 2′-O-Methylation of position 2 of the trypanosome spliced leader cap 4 is mediated by a 48kDa protein related to vaccinia virus VP39. Mol. Biochem. Parasitol. 147:137-139. - PubMed
-
- Bangs, J. D., P. F. Crain, T. Hashizume, J. A. McCloskey, and J. C. Boothroyd. 1992. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J. Biol. Chem. 267:9805-9815. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
