Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination
- PMID: 17606631
- PMCID: PMC2118634
- DOI: 10.1084/jem.20070255
Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination
Abstract
V(D)J recombination and immunoglobulin class switch recombination (CSR) are two somatic rearrangement mechanisms that proceed through the introduction of double-strand breaks (DSBs) in DNA. Although the DNA repair factor XRCC4 is essential for the resolution of DNA DSB during V(D)J recombination, its role in CSR has not been established. To bypass the embryonic lethality of XRCC4 deletion in mice, we developed a conditional XRCC4 knockout (KO) using LoxP-flanked XRCC4 cDNA lentiviral transgenesis. B lymphocyte restricted deletion of XRCC4 in these mice lead to an average two-fold reduction in CSR in vivo and in vitro. Our results connect XRCC4 and the nonhomologous end joining DNA repair pathway to CSR while reflecting the possible use of an alternative pathway in the repair of CSR DSB in the absence of XRCC4. In addition, this new conditional KO approach should be useful in studying other lethal mutations in mice.
Figures
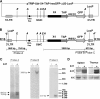
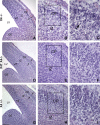

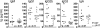



References
-
- de Villartay, J.P., A. Fischer, and A. Durandy. 2003. The mechanisms of immune diversification and their disorders. Nat. Rev. Immunol. 3:962–972. - PubMed
-
- Revy, P., D. Buck, F. le Deist, and J.P. de Villartay. 2005. The repair of DNA damages/modifications during the maturation of the immune system: lessons from human primary immunodeficiency disorders and animal models. Adv. Immunol. 87:237–295. - PubMed
-
- Buck, D., L. Malivert, R. de Chasseval, A. Barraud, M.C. Fondaneche, O. Sanal, A. Plebani, J.L. Stephan, M. Hufnagel, F. le Deist, et al. 2006. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 124:287–299. - PubMed
-
- Callebaut, I., L. Malivert, A. Fischer, J.P. Mornon, P. Revy, and J.P. de Villartay. 2006. Cernunnos interacts with the XRCC4 x DNA-ligase IV complex and is homologous to the yeast nonhomologous end-joining factor Nej1. J. Biol. Chem. 281:13857–13860. - PubMed
-
- Ahnesorg, P., P. Smith, and S.P. Jackson. 2006. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 124:301–313. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

