A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons
- PMID: 17606642
- PMCID: PMC1899473
- DOI: 10.1101/gad.1558107
A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons
Abstract
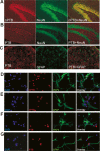
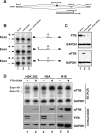
Many metazoan gene transcripts exhibit neuron-specific splicing patterns, but the developmental control of these splicing events is poorly understood. We show that the splicing of a large group of exons is reprogrammed during neuronal development by a switch in expression between two highly similar polypyrimidine tract-binding proteins, PTB and nPTB (neural PTB). PTB is a well-studied regulator of alternative splicing, but nPTB is a closely related paralog whose functional relationship to PTB is unknown. In the brain, nPTB protein is specifically expressed in post-mitotic neurons, whereas PTB is restricted to neuronal precursor cells (NPC), glia, and other nonneuronal cells. Interestingly, nPTB mRNA transcripts are found in NPCs and other nonneuronal cells, but in these cells nPTB protein expression is repressed. This repression is due in part to PTB-induced alternative splicing of nPTB mRNA, leading to nonsense-mediated decay (NMD). However, we find that even properly spliced mRNA fails to express nPTB protein when PTB is present, indicating contributions from additional post-transcriptional mechanisms. The PTB-controlled repression of nPTB results in a mutually exclusive pattern of expression in the brain, where the loss of PTB in maturing neurons allows the synthesis of nPTB in these cells. To examine the consequences of this switch, we used splicing-sensitive microarrays to identify different sets of exons regulated by PTB, nPTB, or both proteins. During neuronal differentiation, the splicing of these exon sets is altered as predicted from the observed changes in PTB and nPTB expression. These data show that the post-transcriptional switch from PTB to nPTB controls a widespread alternative splicing program during neuronal development.
Figures





Comment in
-
PTB/nPTB switch: a post-transcriptional mechanism for programming neuronal differentiation.Genes Dev. 2007 Jul 1;21(13):1573-7. doi: 10.1101/gad.1575607. Genes Dev. 2007. PMID: 17606635 Review. No abstract available.
References
-
- Ahlemeyer B., Baumgart-Vogt E., Baumgart-Vogt E. Optimized protocols for the simultaneous preparation of primary neuronal cultures of the neocortex, hippocampus and cerebellum from individual newborn (P0.5) C57Bl/6J mice. J. Neurosci. Methods. 2005;149:110–120. - PubMed
-
- Ballas N., Grunseich C., Lu D.D., Speh J.C., Mandel G., Grunseich C., Lu D.D., Speh J.C., Mandel G., Lu D.D., Speh J.C., Mandel G., Speh J.C., Mandel G., Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. - PubMed
-
- Bejerano G., Pheasant M., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Pheasant M., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Stephen S., Kent W.J., Mattick J.S., Haussler D., Kent W.J., Mattick J.S., Haussler D., Mattick J.S., Haussler D., Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
