Splicing remodels messenger ribonucleoprotein architecture via eIF4A3-dependent and -independent recruitment of exon junction complex components
- PMID: 17606899
- PMCID: PMC1913901
- DOI: 10.1073/pnas.0704946104
Splicing remodels messenger ribonucleoprotein architecture via eIF4A3-dependent and -independent recruitment of exon junction complex components
Abstract
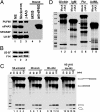
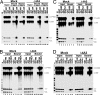
Pre-mRNA splicing not only removes introns and joins exons to generate spliced mRNA but also results in remodeling of the spliced messenger ribonucleoprotein, influencing various downstream events. This remodeling includes the loading of an exon-exon junction complex (EJC). It is unclear how the spliceosome recruits the EJC onto the mRNA and whether EJC formation or EJC components are required for pre-mRNA splicing. Here we immunodepleted the EJC core component eIF4A3 from HeLa cell nuclear extract and found that eIF4A3 is dispensable for pre-mRNA splicing in vitro. However, eIF4A3 is required for the splicing-dependent loading of the Y14/Magoh heterodimer onto mRNA, and this activity of human eIF4A3 is also present in the Drosophila ortholog. Surprisingly, the loading of six other EJC components was not affected by eIF4A3 depletion, suggesting that their binding to mRNA involves different or redundant pathways. Finally, we found that the assembly of the EJC onto mRNA occurs at the late stages of the splicing reaction and requires the second-step splicing and mRNA-release factor HRH1/hPrp22. The EJC-dependent and -independent recruitment of RNA-binding proteins onto mRNA suggests a role for the EJC in messenger ribonucleoprotein remodeling involving interactions with other proteins already bound to the pre-mRNA, which has implications for nonsense-mediated mRNA decay and other mRNA transactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
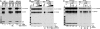

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

