The calpains: modular designs and functional diversity
- PMID: 17608959
- PMCID: PMC2394746
- DOI: 10.1186/gb-2007-8-6-218
The calpains: modular designs and functional diversity
Abstract
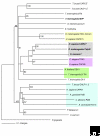
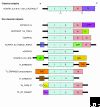
The calpain family is named for the calcium dependence of the papain-like, thiol protease activity of the well-studied ubiquitous vertebrate enzymes calpain-1 (mu-calpain) and calpain-2 (m-calpain). Proteins showing sequence relatedness to the catalytic core domains of these enzymes are included in this ancient and diverse eukaryotic protein family. Calpains are examples of highly modular organization, with several varieties of amino-terminal or carboxy-terminal modules flanking a conserved core. Acquisition of the penta-EF-hand module involved in calcium binding (and the formation of heterodimers for some calpains) seems to be a relatively late event in calpain evolution. Several alternative mechanisms for binding calcium and associating with membranes/phospholipids are found throughout the family. The gene family is expanded in mammals, trypanosomes and ciliates, with up to 26 members in Tetrahymena, for example; in striking contrast to this, only a single calpain gene is present in many other protozoa and in plants. The many isoforms of calpain and their multiple splice variants complicate the discussion and analysis of the family, and challenge researchers to ascertain the relationships between calpain gene sequences, protein isoforms and their distinct or overlapping functions. In mammals and plants it is clear that a calpain plays an essential role in development. There is increasing evidence that ubiquitous calpains participate in a variety of signal transduction pathways and function in important cellular processes of life and death. In contrast to relatively promiscuous degradative proteases, calpains cleave only a restricted set of protein substrates and use complex substrate-recognition mechanisms, involving primary and secondary structural features of target proteins. The detailed physiological significance of both proteolytically active calpains and those lacking key catalytic residues requires further study.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

