Role of the cytoplasmic tail domains of Bunyamwera orthobunyavirus glycoproteins Gn and Gc in virus assembly and morphogenesis
- PMID: 17609275
- PMCID: PMC2045389
- DOI: 10.1128/JVI.00573-07
Role of the cytoplasmic tail domains of Bunyamwera orthobunyavirus glycoproteins Gn and Gc in virus assembly and morphogenesis
Abstract
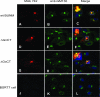
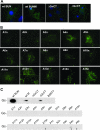
The M RNA genome segment of Bunyamwera virus (BUNV), the prototype of the Bunyaviridae family, encodes a precursor polyprotein that is proteolytically cleaved to yield two structural proteins, Gn and Gc, and a nonstructural protein called NSm. Gn and Gc are type I integral transmembrane glycoproteins. The Gn protein contains a predicted cytoplasmic tail (CT) of 78 residues, and Gc has a shorter CT of 25 residues. Little is known about the role of the Gn and Gc CT domains in the virus replication cycle. We generated a series of mutant glycoprotein precursor constructs containing either deletions or alanine substitutions in the CT domains of Gn and Gc. We examined the effects of these mutations on glycoprotein maturation, cell surface expression, and low pH-induced syncytium formation. In addition, the effects of these mutations were also assessed using a reverse genetics-based virus assembly assay and a virus rescue system. Our results show that the CT domains of both Gn and Gc play crucial roles in BUNV-mediated membrane fusion, virus assembly, and morphogenesis.
Figures



Similar articles
-
Bunyamwera orthobunyavirus glycoprotein precursor is processed by cellular signal peptidase and signal peptide peptidase.Proc Natl Acad Sci U S A. 2016 Aug 2;113(31):8825-30. doi: 10.1073/pnas.1603364113. Epub 2016 Jul 20. Proc Natl Acad Sci U S A. 2016. PMID: 27439867 Free PMC article.
-
Requirement of the N-terminal region of orthobunyavirus nonstructural protein NSm for virus assembly and morphogenesis.J Virol. 2006 Aug;80(16):8089-99. doi: 10.1128/JVI.00579-06. J Virol. 2006. PMID: 16873265 Free PMC article.
-
Mapping the Golgi targeting and retention signal of Bunyamwera virus glycoproteins.J Virol. 2004 Oct;78(19):10793-802. doi: 10.1128/JVI.78.19.10793-10802.2004. J Virol. 2004. PMID: 15367646 Free PMC article.
-
The Role of Phlebovirus Glycoproteins in Viral Entry, Assembly and Release.Viruses. 2016 Jul 21;8(7):202. doi: 10.3390/v8070202. Viruses. 2016. PMID: 27455305 Free PMC article. Review.
-
Hantavirus Gn and Gc envelope glycoproteins: key structural units for virus cell entry and virus assembly.Viruses. 2014 Apr 21;6(4):1801-22. doi: 10.3390/v6041801. Viruses. 2014. PMID: 24755564 Free PMC article. Review.
Cited by
-
Systems to establish bunyavirus genome replication in the absence of transcription.J Virol. 2013 Jul;87(14):8205-12. doi: 10.1128/JVI.00371-13. Epub 2013 May 22. J Virol. 2013. PMID: 23698297 Free PMC article.
-
Throw out the Map: Neuropathogenesis of the Globally Expanding California Serogroup of Orthobunyaviruses.Viruses. 2019 Aug 29;11(9):794. doi: 10.3390/v11090794. Viruses. 2019. PMID: 31470541 Free PMC article. Review.
-
Circulation of diverse genotypes of Tahyna virus in Xinjiang, People's Republic of China.Am J Trop Med Hyg. 2011 Sep;85(3):442-5. doi: 10.4269/ajtmh.2011.10-0368. Am J Trop Med Hyg. 2011. PMID: 21896801 Free PMC article.
-
Functional analysis of the Bunyamwera orthobunyavirus Gc glycoprotein.J Gen Virol. 2009 Oct;90(Pt 10):2483-2492. doi: 10.1099/vir.0.013540-0. Epub 2009 Jul 1. J Gen Virol. 2009. PMID: 19570952 Free PMC article.
-
The Hantavirus Glycoprotein G1 Tail Contains Dual CCHC-type Classical Zinc Fingers.J Biol Chem. 2009 Mar 27;284(13):8654-60. doi: 10.1074/jbc.M808081200. Epub 2009 Jan 29. J Biol Chem. 2009. PMID: 19179334 Free PMC article.
References
-
- Anderson, G. W., and J. F. Smith. 1987. Immunoelectron microscopy of Rift Valley fever viral morphogenesis in primary rat hepatocytes. Virology 161:91-100. - PubMed
-
- Arikawa, J., I. Takashima, and N. Hashimoto. 1985. Cell fusion by haemorrhagic fever with renal syndrome (HFRS) viruses and its application for titration of virus infectivity and neutralizing antibody. Arch. Virol. 86:303-313. - PubMed
-
- Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. - PMC - PubMed
-
- Celma, C. C. P., J. M. Manrique, J. L. Affranchino, E. Hunter, and S. A. Gonzalez. 2001. Domains in the simian immunodeficiency virus gp41 cytoplasmic tail required for envelope incorporation into particles. Virology 283:253-261. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

