Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006
- PMID: 17609280
- PMCID: PMC2045401
- DOI: 10.1128/JVI.00674-07
Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006
Abstract
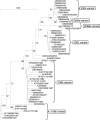
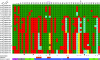
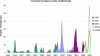
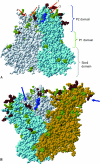
Noroviruses are the causative agents of the majority of viral gastroenteritis outbreaks in humans. During the past 15 years, noroviruses of genotype GGII.4 have caused four epidemic seasons of viral gastroenteritis, during which four novel variants (termed epidemic variants) emerged and displaced the resident viruses. In order to understand the mechanisms and biological advantages of these epidemic variants, we studied the genetic changes in the capsid proteins of GGII.4 strains over this period. A representative sample was drawn from 574 GGII.4 outbreak strains collected over 15 years of systematic surveillance in The Netherlands, and capsid genes were sequenced for a total of 26 strains. The three-dimensional structure was predicted by homology modeling, using the Norwalk virus (Hu/NoV/GGI.1/Norwalk/1968/US) capsid as a reference. The highly significant preferential accumulation and fixation of mutations (nucleotide and amino acid) in the protruding part of the capsid protein provided strong evidence for the occurrence of genetic drift and selection. Although subsequent new epidemic variants differed by up to 25 amino acid mutations, consistent changes were observed in only five positions. Phylogenetic analyses showed that each variant descended from its chronologic predecessor, with the exception of the 2006b variant, which is more closely related to the 2002 variant than to the 2004 variant. The consistent association between the observed genetic findings and changes in epidemiology leads to the conclusion that population immunity plays a role in the epochal evolution of GGII.4 norovirus strains.
Figures


References
-
- Ando, T., J. S. Noel, and R. L. Fankhauser. 2000. Genetic classification of Norwalk-like viruses. J. Infect. Dis. 181(Suppl. 2):S336-S348. - PubMed
-
- Blanton, L. H., S. M. Adams, R. S. Beard, G. Wei, S. N. Bulens, M. A. Widdowson, R. I. Glass, and S. S. Monroe. 2006. Molecular and epidemiologic trends of caliciviruses associated with outbreaks of acute gastroenteritis in the United States, 2000-2004. J. Infect. Dis. 193:413-421. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

