Real-time PCR assay for rapid detection and analysis of PfCRT haplotypes of chloroquine-resistant Plasmodium falciparum isolates from India
- PMID: 17609321
- PMCID: PMC2045286
- DOI: 10.1128/JCM.02291-06
Real-time PCR assay for rapid detection and analysis of PfCRT haplotypes of chloroquine-resistant Plasmodium falciparum isolates from India
Abstract
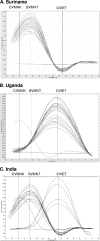
Chloroquine-resistant Plasmodium falciparum (CRPF) malaria isolates in Southeast Asia and sub-Saharan Africa share the same Plasmodium falciparum chloroquine resistance transporter (PfCRT) haplotype (CVIET; amino acids 72 to 76). It is believed that CRPF malaria emerged in Southeast Asia and spread to sub-Saharan Africa via the Indian subcontinent. Based on this assumption, we hypothesized that CRPF isolates in India should possess the same drug resistance haplotype (PfCRT haplotype CVIET) as P. falciparum isolates in Southeast Asia and Africa and that the prevalence of CRPF may be higher and more widespread in India than appreciated. To test this postulate, we utilized a standardized real-time PCR assay to assess the prevalence and distribution of PfCRT haplotypes in P. falciparum isolates (n = 406) collected from Western, Central, and Eastern states in India and compared them to isolates from South America and Africa. Based on the proportion of isolates possessing the molecular marker K76T, the prevalence of chloroquine resistance was high in all five regions of India studied (91%), as well as in Uganda (98%) and Suriname (100%). All isolates from Suriname contained the chloroquine-resistant SVMNT haplotype typical of South American isolates, and 98% of isolates from Uganda possessed the chloroquine-resistant CVIET haplotype characteristic of Southeast Asian/African strains. However, of 246 P. falciparum isolates from across India that contained the molecular marker for chloroquine resistance, 81% contained the SVMNT haplotype. In conclusion, the prevalence of CRPF malaria was high in geographically dispersed regions of India, and the primary haplotype observed, SVMNT, did not support a presumed geographic spread from contiguous Southeast Asia.
Figures
Similar articles
-
Population genetic analyses of Plasmodium falciparum chloroquine receptor transporter gene haplotypes reveal the evolutionary history of chloroquine-resistant malaria in India.Int J Parasitol. 2011 Jun;41(7):705-9. doi: 10.1016/j.ijpara.2011.03.002. Epub 2011 Apr 9. Int J Parasitol. 2011. PMID: 21447338
-
Widespread occurrence of the Plasmodium falciparum chloroquine resistance transporter (Pfcrt) gene haplotype SVMNT in P. falciparum malaria in India.Am J Trop Med Hyg. 2004 Mar;70(3):256-9. Am J Trop Med Hyg. 2004. PMID: 15031513
-
Association of pfcrt but not pfmdr1 alleles with chloroquine resistance in Iranian isolates of Plasmodium falciparum.Am J Trop Med Hyg. 2008 Apr;78(4):633-40. Am J Trop Med Hyg. 2008. PMID: 18385361
-
pfcrt is more than the Plasmodium falciparum chloroquine resistance gene: a functional and evolutionary perspective.Acta Trop. 2005 Jun;94(3):170-80. doi: 10.1016/j.actatropica.2005.04.004. Acta Trop. 2005. PMID: 15866507 Review.
-
Spread and evolution of Plasmodium falciparum drug resistance.Parasitol Int. 2009 Sep;58(3):201-9. doi: 10.1016/j.parint.2009.04.004. Epub 2009 Apr 23. Parasitol Int. 2009. PMID: 19393762 Review.
Cited by
-
Factors associated with regional bias of pfcrt (plasmodium falciparum chloroquine resistance transporter) haplotypes in Nepal.Southeast Asian J Trop Med Public Health. 2011 Jan;42(1):1-8. Southeast Asian J Trop Med Public Health. 2011. PMID: 21323158 Free PMC article.
-
Mutant pfcrt "SVMNT" haplotype and wild type pfmdr1 "N86" are endemic in Plasmodium vivax dominated areas of India under high chloroquine exposure.Malar J. 2012 Jan 11;11:16. doi: 10.1186/1475-2875-11-16. Malar J. 2012. PMID: 22236376 Free PMC article.
-
Prevalence of mutations associated with antimalarial drugs in Plasmodium falciparum isolates prior to the introduction of sulphadoxine-pyrimethamine as first-line treatment in Iran.Malar J. 2007 Nov 13;6:148. doi: 10.1186/1475-2875-6-148. Malar J. 2007. PMID: 17999755 Free PMC article.
-
Detection of Plasmodium vivax by nested PCR and real-time PCR.Korean J Parasitol. 2010 Jun;48(2):99-103. doi: 10.3347/kjp.2010.48.2.99. Epub 2010 Jun 17. Korean J Parasitol. 2010. PMID: 20585524 Free PMC article.
-
Molecular assessment of Plasmodium falciparum resistance to antimalarial drugs in Papua New Guinea using an extended ligase detection reaction fluorescent microsphere assay.Antimicrob Agents Chemother. 2011 Feb;55(2):798-805. doi: 10.1128/AAC.00939-10. Epub 2010 Nov 15. Antimicrob Agents Chemother. 2011. PMID: 21078925 Free PMC article.
References
-
- Anderson, T. J., B. Haubold, J. T. Williams, J. G. Estrada-Franco, L. Richardson, R. Mollinedo, M. Bockarie, J. Mokili, S. Mharakurwa, N. French, J. Whitworth, I. D. Velez, A. H. Brockman, F. Nosten, M. U. Ferreira, and K. P. Day. 2000. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol. Biol. Evol. 17:1467-1482. - PubMed
-
- Attaran, A., K. I. Barnes, R. Bate, F. Binka, U. d'Alessandro, C. I. Fanello, L. Garrett, T. K. Mutabingwa, D. Roberts, C. H. Sibley, A. Talisuna, J. P. Van Geertruyden, and W. M. Watkins. 2006. The World Bank: false financial and statistical accounts and medical malpractice in malaria treatment. Lancet 368:247-252. - PubMed
-
- Bray, P. G., R. E. Martin, L. Tilley, S. A. Ward, K. Kirk, and D. A. Fidock. 2005. Defining the role of PfCRT in Plasmodium falciparum chloroquine resistance. Mol. Microbiol. 56:323-333. - PubMed
-
- Carlton, J. M., D. A. Fidock, A. Djimde, C. V. Plowe, and T. E. Wellems. 2001. Conservation of a novel vacuolar transporter in Plasmodium species and its central role in chloroquine resistance of P. falciparum. Curr. Opin. Microbiol. 4:415-420. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources