Laboratory evolution of one disulfide isomerase to resemble another
- PMID: 17609373
- PMCID: PMC1906722
- DOI: 10.1073/pnas.0704692104
Laboratory evolution of one disulfide isomerase to resemble another
Abstract
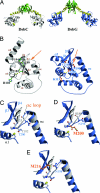
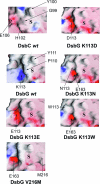
It is often difficult to determine which of the sequence and structural differences between divergent members of multigene families are functionally important. Here we use a laboratory evolution approach to determine functionally important structural differences between two distantly related disulfide isomerases, DsbC and DsbG from Escherichia coli. Surprisingly, we found single amino acid substitutions in DsbG that were able to complement dsbC in vivo and have more DsbC-like isomerase activity in vitro. Crystal structures of the three strongest point mutants, DsbG K113E, DsbG V216M, and DsbG T200M, reveal changes in highly surface-exposed regions that cause DsbG to more closely resemble the distantly related DsbC. In this case, laboratory evolution appears to have taken a direct route to allow one protein family member to complement another, with single substitutions apparently bypassing much of the need for multiple changes that took place over approximately 0.5 billion years of evolution. Our findings suggest that, for these two proteins at least, regions important in determining functional differences may represent only a tiny fraction of the overall protein structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

