The design and characterization of two proteins with 88% sequence identity but different structure and function
- PMID: 17609385
- PMCID: PMC1906725
- DOI: 10.1073/pnas.0700922104
The design and characterization of two proteins with 88% sequence identity but different structure and function
Abstract
To identify a simplified code for conformational switching, we have redesigned two natural proteins to have 88% sequence identity but different tertiary structures: a 3-alpha helix fold and an alpha/beta fold. We describe the design of these homologous heteromorphic proteins, their structural properties as determined by NMR, their conformational stabilities, and their affinities for their respective ligands: IgG and serum albumin. Each of these proteins is completely folded at 25 degrees C, is monomeric, and retains the native binding activity. The complete binding epitope for both ligands is encoded within each of the proteins. The IgG-binding epitope is functional only in the alpha/beta fold, and the albumin-binding epitope is functional only in the 3-alpha fold. These results demonstrate that two monomeric folds and two different functions can be encoded with only 12% of the amino acids in a protein (7 of 56). The fact that 49 aa in these proteins are compatible with both folds shows that the essential information determining a fold can be highly concentrated in a few amino acids and that a very limited subset of interactions in the protein can tip the balance from one monomer fold to another. This delicate balance helps explain why protein structure prediction is so challenging. Furthermore, because a few mutations can result in both new conformation and new function, the evolution of new folds driven by natural selection for alternative functions may be much more probable than previously recognized.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

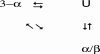





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

