TRPM5, a taste-signaling transient receptor potential ion-channel, is a ubiquitous signaling component in chemosensory cells
- PMID: 17610722
- PMCID: PMC1931605
- DOI: 10.1186/1471-2202-8-49
TRPM5, a taste-signaling transient receptor potential ion-channel, is a ubiquitous signaling component in chemosensory cells
Abstract
Background: A growing number of TRP channels have been identified as key players in the sensation of smell, temperature, mechanical forces and taste. TRPM5 is known to be abundantly expressed in taste receptor cells where it participates in sweet, amino acid and bitter perception. A role of TRPM5 in other sensory systems, however, has not been studied so far.
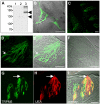
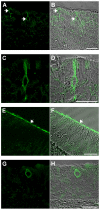
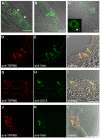
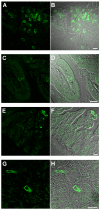
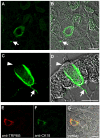
Results: Here, we systematically investigated the expression of TRPM5 in rat and mouse tissues. Apart from taste buds, where we found TRPM5 to be predominantly localized on the basolateral surface of taste receptor cells, TRPM5 immunoreactivity was seen in other chemosensory organs - the main olfactory epithelium and the vomeronasal organ. Most strikingly, we found solitary TRPM5-enriched epithelial cells in all parts of the respiratory and gastrointestinal tract. Based on their tissue distribution, the low cell density, morphological features and co-immunostaining with different epithelial markers, we identified these cells as brush cells (also known as tuft, fibrillovesicular, multivesicular or caveolated cells). In terms of morphological characteristics, brush cells resemble taste receptor cells, while their origin and biological role are still under intensive debate.
Conclusion: We consider TRPM5 to be an intrinsic signaling component of mammalian chemosensory organs, and provide evidence for brush cells being an important cellular correlate in the periphery.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

