The ZFHX1A gene is differentially autoregulated by its isoforms
- PMID: 17610840
- PMCID: PMC2770808
- DOI: 10.1016/j.bbrc.2007.06.088
The ZFHX1A gene is differentially autoregulated by its isoforms
Abstract
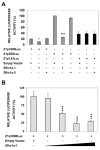
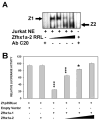
The Zfhx1a gene expresses two different isoforms; the full length Zfhx1a-1 and a truncated isoform termed Zfhx1a-2 lacking the first exon. Deletion analysis of the Zfhx1a-1 promoter localized cell-specific repressors, and a proximal G-string that is critically required for transactivation. Transfection of Zfhx1a-1 cDNA, but not Zfhx1a-2, downregulates Zfhx1a-1 promoter activity. Mutation of an E2-box disrupted the binding of both Zfhx1a isoforms. Consistent with this, transfected Zfhx1a-1 does not regulate the transcriptional activity of the E-box mutated Zfhx1a-1 promoter. Competitive EMSAs and transfection assays show that Zfhx1a-2 can function as a dominant negative isoform since it is able to compete and displace Zfhx1a-1 from its binding site and overcome Zfhx1a-1 induced repression of the Zfhx1a-1 promoter in cells. Hence, the Zfhx1a-1 gene is autoregulated in part by negative feedback on its own promoter which is, in turn, modified by the availability of the negative dominant isoform Zfhx1a-2.
Figures


Similar articles
-
Estrogen enhances gonadotropin-releasing hormone-stimulated transcription of the luteinizing hormone subunit promoters via altered expression of stimulatory and suppressive transcription factors.Endocrinology. 2007 Dec;148(12):6083-91. doi: 10.1210/en.2007-0407. Epub 2007 Sep 6. Endocrinology. 2007. PMID: 17823254
-
The zinc finger transcription factor ZFHX1A is linked to cell proliferation by Rb-E2F1.Biochem J. 2007 Nov 15;408(1):79-85. doi: 10.1042/BJ20070344. Biochem J. 2007. PMID: 17655524 Free PMC article.
-
PITX2 isoform-specific regulation of atrial natriuretic factor expression: synergism and repression with Nkx2.5.J Biol Chem. 2003 Jun 20;278(25):22437-45. doi: 10.1074/jbc.M210163200. Epub 2003 Apr 11. J Biol Chem. 2003. PMID: 12692125
-
Sp proteins and Phox2b regulate the expression of the human Phox2a gene.J Neurosci. 2001 Sep 15;21(18):7037-45. doi: 10.1523/JNEUROSCI.21-18-07037.2001. J Neurosci. 2001. PMID: 11549713 Free PMC article.
-
The role of the ZEB family of transcription factors in development and disease.Cell Mol Life Sci. 2009 Mar;66(5):773-87. doi: 10.1007/s00018-008-8465-8. Cell Mol Life Sci. 2009. PMID: 19011757 Free PMC article. Review.
Cited by
-
CCN6 (WISP3) decreases ZEB1-mediated EMT and invasion by attenuation of IGF-1 receptor signaling in breast cancer.J Cell Sci. 2011 May 15;124(Pt 10):1752-8. doi: 10.1242/jcs.084194. Epub 2011 Apr 26. J Cell Sci. 2011. PMID: 21525039 Free PMC article.
-
Comparative Genomics of the BDNF Gene, Non-Canonical Modes of Transcriptional Regulation, and Neurological Disease.Mol Neurobiol. 2021 Jun;58(6):2851-2861. doi: 10.1007/s12035-021-02306-z. Epub 2021 Jan 30. Mol Neurobiol. 2021. PMID: 33517560 Review.
-
Phosphorylation Regulates Functions of ZEB1 Transcription Factor.J Cell Physiol. 2016 Oct;231(10):2205-17. doi: 10.1002/jcp.25338. Epub 2016 Mar 10. J Cell Physiol. 2016. PMID: 26868487 Free PMC article.
-
Transcriptional Characterization of Porcine Leptin and Leptin Receptor Genes.PLoS One. 2013 Jun 18;8(6):e66398. doi: 10.1371/journal.pone.0066398. Print 2013. PLoS One. 2013. PMID: 23824082 Free PMC article.
-
Expression of the ZEB1 (deltaEF1) transcription factor in human: additional insights.Mol Cell Biochem. 2008 Nov;318(1-2):89-99. doi: 10.1007/s11010-008-9860-z. Epub 2008 Jul 12. Mol Cell Biochem. 2008. PMID: 18622689
References
-
- Darling D, Gaur N, Zhu B. A zinc finger homeodomain transcription factor binds specific thyroid hormone response elements. Mol Cell Endocrinol. 1998;139:25–35. - PubMed
-
- Williams T, Moolten D, Burlein J, Romano J, Bhaerman R, Godillot A, Mellon M, Rauscher F, 3rd, Kant J. Identification of a zinc finger protein that inhibits IL-2 gene expression. Science. 1991;254:1791–1794. - PubMed
-
- Takagi T, Moribe H, Kondoh H, Higashi Y. DeltaEF1, a zinc finger and homeodomain transcription factor, is required for skeleton patterning in multiple lineages. Development. 1998;125:21–31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

