Alkyne substrate interaction within the nitrogenase MoFe protein
- PMID: 17610955
- PMCID: PMC2711850
- DOI: 10.1016/j.jinorgbio.2007.05.007
Alkyne substrate interaction within the nitrogenase MoFe protein
Abstract
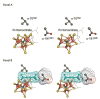
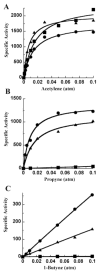
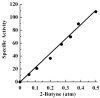
Nitrogenase catalyzes the biological reduction of N(2) to ammonia (nitrogen fixation), as well as the two-electron reduction of the non-physiological alkyne substrate acetylene (HC triple bond CH). A complex metallo-organic species called FeMo-cofactor provides the site of substrate reduction within the MoFe protein, but exactly where and how substrates interact with FeMo-cofactor remains unknown. Recent results have shown that the MoFe protein alpha-70(Val) residue, whose side chain approaches one Fe-S face of FeMo-cofactor, plays a significant role in defining substrate access to the active site. For example, substitution of alpha-70(Val) by alanine results in an increased capacity for the reduction of the larger alkyne propyne (HC triple bond C-CH(3)), whereas, substitution by isoleucine at this position nearly eliminates the capacity for the reduction of acetylene. These and complementary spectroscopic studies led us to propose that binding of short chain alkynes occurs with side-on binding to Fe atom 6 within FeMo-cofactor. In the present work, the alpha-70(Val) residue was substituted by glycine and this MoFe protein variant shows an increased capacity for reduction of the terminal alkyne, 1-butyne (HC triple bond C-CH(2)-CH(3)). This protein shows no detectable reduction of the internal alkyne 2-butyne (H(3)C-C triple bond C-CH(3)). In contrast, substitution of the nearby alpha-191(Gln) residue by alanine, in combination with the alpha-70(Ala) substitution, does result in significant reduction of 2-butyne, with the exclusive product being 2-cis-butene. These results indicate that the reduction of alkynes by nitrogenases involves side-on binding of the alkyne to Fe6 within FeMo-cofactor, and that a terminal acidic proton is not required for reduction. The successful design of amino acid substitutions that permit the targeted accommodation of an alkyne that otherwise is not a nitrogenase substrate provides evidence to support the current model for alkyne interaction within the nitrogenase MoFe protein.
Figures


Similar articles
-
Localization of a substrate binding site on the FeMo-cofactor in nitrogenase: trapping propargyl alcohol with an alpha-70-substituted MoFe protein.Biochemistry. 2003 Aug 5;42(30):9102-9. doi: 10.1021/bi034595x. Biochemistry. 2003. PMID: 12885243
-
Role of the MoFe protein alpha-subunit histidine-195 residue in FeMo-cofactor binding and nitrogenase catalysis.Biochemistry. 1995 Mar 7;34(9):2798-808. doi: 10.1021/bi00009a008. Biochemistry. 1995. PMID: 7893691
-
Substrate interaction at an iron-sulfur face of the FeMo-cofactor during nitrogenase catalysis.J Biol Chem. 2004 Dec 17;279(51):53621-4. doi: 10.1074/jbc.M410247200. Epub 2004 Oct 1. J Biol Chem. 2004. PMID: 15465817
-
Breaking the N2 triple bond: insights into the nitrogenase mechanism.Dalton Trans. 2006 May 21;(19):2277-84. doi: 10.1039/b517633f. Epub 2006 Apr 11. Dalton Trans. 2006. PMID: 16688314 Review.
-
Nitrogenase structure and function: a biochemical-genetic perspective.Annu Rev Microbiol. 1995;49:335-66. doi: 10.1146/annurev.mi.49.100195.002003. Annu Rev Microbiol. 1995. PMID: 8561464 Review.
Cited by
-
Biological nitrogen fixation in theory, practice, and reality: a perspective on the molybdenum nitrogenase system.FEBS Lett. 2023 Jan;597(1):45-58. doi: 10.1002/1873-3468.14534. Epub 2022 Nov 28. FEBS Lett. 2023. PMID: 36344435 Free PMC article. Review.
-
Insights into substrate binding at FeMo-cofactor in nitrogenase from the structure of an alpha-70(Ile) MoFe protein variant.J Inorg Biochem. 2010 Apr;104(4):385-9. doi: 10.1016/j.jinorgbio.2009.11.009. Epub 2009 Nov 26. J Inorg Biochem. 2010. PMID: 20022118 Free PMC article.
-
Ligand-Based Control of Single-Site vs. Multi-Site Reactivity by a Trichromium Cluster.Angew Chem Int Ed Engl. 2019 Apr 16;58(17):5687-5691. doi: 10.1002/anie.201901599. Epub 2019 Mar 27. Angew Chem Int Ed Engl. 2019. PMID: 30828957 Free PMC article.
-
Nitrogenase: a draft mechanism.Acc Chem Res. 2013 Feb 19;46(2):587-95. doi: 10.1021/ar300267m. Epub 2013 Jan 4. Acc Chem Res. 2013. PMID: 23289741 Free PMC article.
-
EXAFS and NRVS reveal a conformational distortion of the FeMo-cofactor in the MoFe nitrogenase propargyl alcohol complex.J Inorg Biochem. 2012 Jul;112:85-92. doi: 10.1016/j.jinorgbio.2012.02.004. Epub 2012 Feb 15. J Inorg Biochem. 2012. PMID: 22564272 Free PMC article.
References
-
- Burgess BK, Lowe DJ. Chem Rev. 1996;96:2983–3011. - PubMed
-
- Dos Santos PC, Igarashi RY, Lee HI, Hoffman BM, Seefeldt LC, Dean DR. Acc Chem Res. 2005;38:208–214. - PubMed
-
- Howard JB, Rees DC. Annu Rev Biochem. 1994;63:235–24. - PubMed
-
- Seefeldt LC, Dance I, Dean DR. Biochemistry. 2004;43:1401–1409. - PubMed
-
- Georgiadis MM, Komiya H, Chakrabarti P, Woo D, Kornuc JJ, Rees DC. Science. 1992;257:1653–1659. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

