The roles of Rad16 and Rad26 in repairing repressed and actively transcribed genes in yeast
- PMID: 17611170
- PMCID: PMC2095784
- DOI: 10.1016/j.dnarep.2007.05.005
The roles of Rad16 and Rad26 in repairing repressed and actively transcribed genes in yeast
Abstract
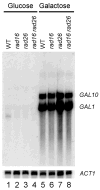
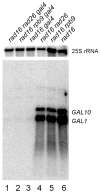
Nucleotide excision repair (NER) is a conserved DNA repair mechanism capable of removing a variety of helix-distorting DNA lesions. Rad26, a member of the Swi2/Snf2 superfamily of proteins, has been shown to be involved in a specialized NER process called transcription coupled NER. Rad16, another member of the same protein superfamily, has been shown to be required for genome-wide NER. Here we show that Rad16 and Rad26 play different roles in repairing repressed and actively transcribed genes in yeast. Rad16 is partially dispensable, and Rad26 plays a significant role in repairing certain regions of the repressed GAL1-10, PHO5 and ADH2 genes, especially in the core DNA of well-positioned nucleosomes. Simultaneous elimination of Rad16 and Rad26 results in no detectable repair in these regions of the repressed genes. Transcriptional induction of the GAL1-10 genes abolishes the role of Rad26, but does not affect the role of Rad16 in repairing the nontranscribed strand of the genes. Interestingly, when the transcription activator Gal4 is eliminated from the cells, Rad16 becomes partially dispensable and Rad26 plays a significant role in repairing both strands of the GAL1-10 genes even under inducing conditions. Our results suggest that Rad16 and Rad26 play different and, to some extent, complementary roles in repairing both strands of repressed genes, although the relative contributions of the two proteins can be different from gene to gene, and from region to region of a gene. However, Rad16 is solely responsible for repairing the nontranscribed strand of actively transcribed genes.
Figures




Similar articles
-
Tfb5 is partially dispensable for Rad26 mediated transcription coupled nucleotide excision repair in yeast.DNA Repair (Amst). 2007 Nov;6(11):1661-9. doi: 10.1016/j.dnarep.2007.06.001. Epub 2007 Jul 20. DNA Repair (Amst). 2007. PMID: 17644494 Free PMC article.
-
Dissecting transcription-coupled and global genomic repair in the chromatin of yeast GAL1-10 genes.J Biol Chem. 2004 Apr 2;279(14):14418-26. doi: 10.1074/jbc.M312004200. Epub 2004 Jan 19. J Biol Chem. 2004. PMID: 14734564 Free PMC article.
-
In UV-irradiated Saccharomyces cerevisiae, overexpression of Swi2/Snf2 family member Rad26 increases transcription-coupled repair and repair of the non-transcribed strand.Mol Microbiol. 2004 Jun;52(6):1653-63. doi: 10.1111/j.1365-2958.2004.04081.x. Mol Microbiol. 2004. PMID: 15186415
-
Transcription coupled nucleotide excision repair in the yeast Saccharomyces cerevisiae: The ambiguous role of Rad26.DNA Repair (Amst). 2015 Dec;36:43-48. doi: 10.1016/j.dnarep.2015.09.006. Epub 2015 Sep 10. DNA Repair (Amst). 2015. PMID: 26429063 Review.
-
Nucleotide excision repair in yeast.Mutat Res. 2000 Jun 30;451(1-2):13-24. doi: 10.1016/s0027-5107(00)00037-3. Mutat Res. 2000. PMID: 10915862 Review.
Cited by
-
RNA polymerase II depletion promotes transcription of alternative mRNA species.BMC Mol Biol. 2016 Aug 30;17(1):20. doi: 10.1186/s12867-016-0074-8. BMC Mol Biol. 2016. PMID: 27578267 Free PMC article.
-
Chromatin structure and DNA damage repair.Epigenetics Chromatin. 2008 Nov 12;1(1):9. doi: 10.1186/1756-8935-1-9. Epigenetics Chromatin. 2008. PMID: 19014481 Free PMC article.
-
Association between DNA repair capacity and body mass index in women.Mutat Res. 2023 Jan-Jun;826:111813. doi: 10.1016/j.mrfmmm.2022.111813. Epub 2022 Dec 31. Mutat Res. 2023. PMID: 36621052 Free PMC article.
-
Arabidopsis RAD16 Homologues Are Involved in UV Tolerance and Growth.Genes (Basel). 2023 Jul 28;14(8):1552. doi: 10.3390/genes14081552. Genes (Basel). 2023. PMID: 37628604 Free PMC article.
-
Organization of DNA damage, excision repair, and mutagenesis in chromatin: A genomic perspective.DNA Repair (Amst). 2019 Sep;81:102645. doi: 10.1016/j.dnarep.2019.102645. Epub 2019 Jul 8. DNA Repair (Amst). 2019. PMID: 31307926 Free PMC article. Review.
References
-
- Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Res Mol Biol. 2005;79:183–235. - PubMed
-
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. ASM Press; Washington D.C: 2006.
-
- Guzder SN, Sung P, Prakash L, Prakash S. The DNA-dependent ATPase activity of yeast nucleotide excision repair factor 4 and its role in DNA damage recognition. J Biol Chem. 1998;273:6292–6296. - PubMed
-
- Verhage R, Zeeman AM, de Groot N, Gleig F, Bang DD, van de Putte P, Brouwer J. The RAD7 and RAD16 genes, which are essential for pyrimidine dimer removal from the silent mating type loci, are also required for repair of the nontranscribed strand of an active gene in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:6135–6142. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

