Genetic diversity among Enterococcus faecalis
- PMID: 17611618
- PMCID: PMC1899230
- DOI: 10.1371/journal.pone.0000582
Genetic diversity among Enterococcus faecalis
Abstract
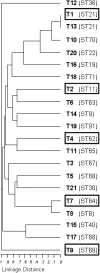
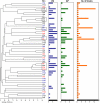
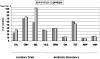
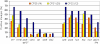
Enterococcus faecalis, a ubiquitous member of mammalian gastrointestinal flora, is a leading cause of nosocomial infections and a growing public health concern. The enterococci responsible for these infections are often resistant to multiple antibiotics and have become notorious for their ability to acquire and disseminate antibiotic resistances. In the current study, we examined genetic relationships among 106 strains of E. faecalis isolated over the past 100 years, including strains identified for their diversity and used historically for serotyping, strains that have been adapted for laboratory use, and isolates from previously described E. faecalis infection outbreaks. This collection also includes isolates first characterized as having novel plasmids, virulence traits, antibiotic resistances, and pathogenicity island (PAI) components. We evaluated variation in factors contributing to pathogenicity, including toxin production, antibiotic resistance, polymorphism in the capsule (cps) operon, pathogenicity island (PAI) gene content, and other accessory factors. This information was correlated with multi-locus sequence typing (MLST) data, which was used to define genetic lineages. Our findings show that virulence and antibiotic resistance traits can be found within many diverse lineages of E. faecalis. However, lineages have emerged that have caused infection outbreaks globally, in which several new antibiotic resistances have entered the species, and in which virulence traits have converged. Comparing genomic hybridization profiles, using a microarray, of strains identified by MLST as spanning the diversity of the species, allowed us to identify the core E. faecalis genome as consisting of an estimated 2057 unique genes.
Conflict of interest statement
Figures



Similar articles
-
Analysis of genetic lineages and their correlation with virulence genes in Enterococcus faecalis clinical isolates from root canal and systemic infections.J Endod. 2013 Jul;39(7):858-64. doi: 10.1016/j.joen.2013.01.009. Epub 2013 Mar 20. J Endod. 2013. PMID: 23791252
-
Capsule locus polymorphism among distinct lineages of Enterococcus faecalis isolated from canals of root-filled teeth with periapical lesions.J Endod. 2012 Jan;38(1):58-61. doi: 10.1016/j.joen.2011.08.002. Epub 2011 Sep 14. J Endod. 2012. PMID: 22152621
-
Emergence of high-risk multidrug-resistant Enterococcus faecalis CC2 (ST181) and CC87 (ST28) causing healthcare-associated infections in India.Infect Genet Evol. 2020 Nov;85:104519. doi: 10.1016/j.meegid.2020.104519. Epub 2020 Aug 30. Infect Genet Evol. 2020. PMID: 32877660
-
Antibiotic resistant enterococci-tales of a drug resistance gene trafficker.Int J Med Microbiol. 2013 Aug;303(6-7):360-79. doi: 10.1016/j.ijmm.2013.03.001. Epub 2013 Mar 8. Int J Med Microbiol. 2013. PMID: 23602510 Review.
-
Enterococcal virulence--pathogenicity island of E. Faecalis.Front Biosci. 2004 Sep 1;9:2335-46. doi: 10.2741/1400. Front Biosci. 2004. PMID: 15353291 Review.
Cited by
-
Pheromone killing of multidrug-resistant Enterococcus faecalis V583 by native commensal strains.Proc Natl Acad Sci U S A. 2015 Jun 9;112(23):7273-8. doi: 10.1073/pnas.1500553112. Epub 2015 May 26. Proc Natl Acad Sci U S A. 2015. PMID: 26039987 Free PMC article.
-
Pan-PCR, a computational method for designing bacterium-typing assays based on whole-genome sequence data.J Clin Microbiol. 2013 Mar;51(3):752-8. doi: 10.1128/JCM.02671-12. Epub 2012 Dec 19. J Clin Microbiol. 2013. PMID: 23254127 Free PMC article.
-
Antimicrobial resistance characterization of Enterococcus faecium, Enterococcus faecalis and Enterococcus hirae isolated from marine coastal recreational waters in the State of São Paulo, Brazil.J Water Health. 2024 Sep;22(9):1628-1640. doi: 10.2166/wh.2024.098. Epub 2024 Aug 23. J Water Health. 2024. PMID: 39340376
-
Reciprocal regulation of cephalosporin resistance in Enterococcus faecalis.mBio. 2011 Nov 1;2(6):e00199-11. doi: 10.1128/mBio.00199-11. Print 2011. mBio. 2011. PMID: 22045988 Free PMC article.
-
Group II Introns Generate Functional Chimeric Relaxase Enzymes with Modified Specificities through Exon Shuffling at Both the RNA and DNA Level.Mol Biol Evol. 2021 Mar 9;38(3):1075-1089. doi: 10.1093/molbev/msaa275. Mol Biol Evol. 2021. PMID: 33118013 Free PMC article.
References
-
- Richards MJ, Edwards JR, Culver DH, Gaynes RP. Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect.Control Hosp.Epidemiol. 2000;21:510–515. - PubMed
-
- Maki DG, Agger WA. Enterococcal bacteremia: clinical features, the risk of endocarditis, and management. Medicine (Baltimore) 1988;67:248–269. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

