Trichostatin A effects on gene expression in the protozoan parasite Entamoeba histolytica
- PMID: 17612405
- PMCID: PMC1940012
- DOI: 10.1186/1471-2164-8-216
Trichostatin A effects on gene expression in the protozoan parasite Entamoeba histolytica
Abstract
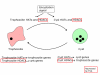
Background: Histone modification regulates chromatin structure and influences gene expression associated with diverse biological functions including cellular differentiation, cancer, maintenance of genome architecture, and pathogen virulence. In Entamoeba, a deep-branching eukaryote, short chain fatty acids (SCFA) affect histone acetylation and parasite development. Additionally, a number of active histone modifying enzymes have been identified in the parasite genome. However, the overall extent of gene regulation tied to histone acetylation is not known.
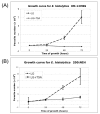
Results: In order to identify the genome-wide effects of histone acetylation in regulating E. histolytica gene expression, we used whole-genome expression profiling of parasites treated with SCFA and Trichostatin A (TSA). Despite significant changes in histone acetylation patterns, exposure of parasites to SCFA resulted in minimal transcriptional changes (11 out of 9,435 genes transcriptionally regulated). In contrast, exposure to TSA, a more specific inhibitor of histone deacetylases, significantly affected transcription of 163 genes (122 genes upregulated and 41 genes downregulated). Genes modulated by TSA were not regulated by treatment with 5-Azacytidine, an inhibitor of DNA-methyltransferase, indicating that in E. histolytica the crosstalk between DNA methylation and histone modification is not substantial. However, the set of genes regulated by TSA overlapped substantially with genes regulated during parasite development: 73/122 genes upregulated by TSA exposure were upregulated in E. histolytica cysts (p-value = 6 x 10(-53)) and 15/41 genes downregulated by TSA exposure were downregulated in E. histolytica cysts (p-value = 3 x 10(-7)).
Conclusion: This work represents the first genome-wide analysis of histone acetylation and its effects on gene expression in E. histolytica. The data indicate that SCFAs, despite their ability to influence histone acetylation, have minimal effects on gene transcription in cultured parasites. In contrast, the effect of TSA on E. histolytica gene expression is more substantial and includes genes involved in the encystation pathway. These observations will allow further dissection of the effects of histone acetylation and the genetic pathways regulating stage conversion in this pathogenic parasite.
Figures
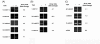

Similar articles
-
Growth of the protozoan parasite Entamoeba histolytica in 5-azacytidine has limited effects on parasite gene expression.BMC Genomics. 2007 Jan 5;8:7. doi: 10.1186/1471-2164-8-7. BMC Genomics. 2007. PMID: 17207281 Free PMC article.
-
Histone acetyltransferases and deacetylase in Entamoeba histolytica.Mol Biochem Parasitol. 2004 Dec;138(2):205-16. doi: 10.1016/j.molbiopara.2004.09.002. Mol Biochem Parasitol. 2004. PMID: 15555732
-
Trichostatin A regulates peroxiredoxin expression and virulence of the parasite Entamoeba histolytica.Mol Biochem Parasitol. 2008 Mar;158(1):82-94. doi: 10.1016/j.molbiopara.2007.11.014. Epub 2007 Dec 4. Mol Biochem Parasitol. 2008. PMID: 18191469
-
Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function.Bioessays. 1995 May;17(5):423-30. doi: 10.1002/bies.950170510. Bioessays. 1995. PMID: 7786288 Review.
-
Proteomics approaches to understand cell biology and virulence of Entamoeba histolytica protozoan parasite.J Proteomics. 2020 Aug 30;226:103897. doi: 10.1016/j.jprot.2020.103897. Epub 2020 Jul 9. J Proteomics. 2020. PMID: 32652218 Review.
Cited by
-
Characterization of an Entamoeba histolytica high-mobility-group box protein induced during intestinal infection.Eukaryot Cell. 2008 Sep;7(9):1565-72. doi: 10.1128/EC.00123-08. Epub 2008 Jul 25. Eukaryot Cell. 2008. PMID: 18658254 Free PMC article.
-
Robust gene silencing mediated by antisense small RNAs in the pathogenic protist Entamoeba histolytica.Nucleic Acids Res. 2013 Nov;41(20):9424-37. doi: 10.1093/nar/gkt717. Epub 2013 Aug 9. Nucleic Acids Res. 2013. PMID: 23935116 Free PMC article.
-
Epigenome mapping highlights chromatin-mediated gene regulation in the protozoan parasite Trichomonas vaginalis.Sci Rep. 2017 Mar 27;7:45365. doi: 10.1038/srep45365. Sci Rep. 2017. PMID: 28345651 Free PMC article.
-
Identification of an Entamoeba histolytica serine-, threonine-, and isoleucine-rich protein with roles in adhesion and cytotoxicity.Eukaryot Cell. 2007 Nov;6(11):2139-46. doi: 10.1128/EC.00174-07. Epub 2007 Sep 7. Eukaryot Cell. 2007. PMID: 17827347 Free PMC article.
-
Targets of the Entamoeba histolytica transcription factor URE3-BP.PLoS Negl Trop Dis. 2008;2(8):e282. doi: 10.1371/journal.pntd.0000282. Epub 2008 Aug 27. PLoS Negl Trop Dis. 2008. PMID: 18846235 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

