Cell-specific microarray profiling experiments reveal a comprehensive picture of gene expression in the C. elegans nervous system
- PMID: 17612406
- PMCID: PMC2323220
- DOI: 10.1186/gb-2007-8-7-r135
Cell-specific microarray profiling experiments reveal a comprehensive picture of gene expression in the C. elegans nervous system
Abstract
Background: With its fully sequenced genome and simple, well-defined nervous system, the nematode Caenorhabditis elegans offers a unique opportunity to correlate gene expression with neuronal differentiation. The lineal origin, cellular morphology and synaptic connectivity of each of the 302 neurons are known. In many instances, specific behaviors can be attributed to particular neurons or circuits. Here we describe microarray-based methods that monitor gene expression in C. elegans neurons and, thereby, link comprehensive profiles of neuronal transcription to key developmental and functional properties of the nervous system.
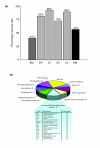
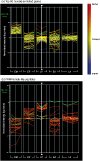
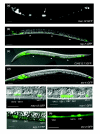
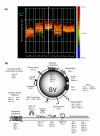
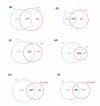
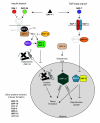
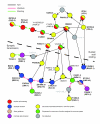
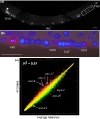
Results: We employed complementary microarray-based strategies to profile gene expression in the embryonic and larval nervous systems. In the MAPCeL (Microarray Profiling C. elegans cells) method, we used fluorescence activated cell sorting (FACS) to isolate GFP-tagged embryonic neurons for microarray analysis. To profile the larval nervous system, we used the mRNA-tagging technique in which an epitope-labeled mRNA binding protein (FLAG-PAB-1) was transgenically expressed in neurons for immunoprecipitation of cell-specific transcripts. These combined approaches identified approximately 2,500 mRNAs that are highly enriched in either the embryonic or larval C. elegans nervous system. These data are validated in part by the detection of gene classes (for example, transcription factors, ion channels, synaptic vesicle components) with established roles in neuronal development or function. Of particular interest are 19 conserved transcripts of unknown function that are also expressed in the mammalian brain. In addition to utilizing these profiling approaches to define stage-specific gene expression, we also applied the mRNA-tagging method to fingerprint a specific neuron type, the A-class group of cholinergic motor neurons, during early larval development. A comparison of these data to a MAPCeL profile of embryonic A-class motor neurons identified genes with common functions in both types of A-class motor neurons as well as transcripts with roles specific to each motor neuron type.
Conclusion: We describe microarray-based strategies for generating expression profiles of embryonic and larval C. elegans neurons. These methods can be applied to particular neurons at specific developmental stages and, therefore, provide an unprecedented opportunity to obtain spatially and temporally defined snapshots of gene expression in a simple model nervous system.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- DK58749/DK/NIDDK NIH HHS/United States
- T32 MH064913/MH/NIMH NIH HHS/United States
- P01 DK58212/DK/NIDDK NIH HHS/United States
- T32 MH64913/MH/NIMH NIH HHS/United States
- T32 HD007502/HD/NICHD NIH HHS/United States
- P60 DK020593/DK/NIDDK NIH HHS/United States
- F31 NS043068/NS/NINDS NIH HHS/United States
- P30 EY08126/EY/NEI NIH HHS/United States
- F31 NS049743/NS/NINDS NIH HHS/United States
- R01 NS026115/NS/NINDS NIH HHS/United States
- T32 HD07502/HD/NICHD NIH HHS/United States
- P30 CA68485/CA/NCI NIH HHS/United States
- U01 HG004263/HG/NHGRI NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- P30 HD015052/HD/NICHD NIH HHS/United States
- V01 HG004263/HG/NHGRI NIH HHS/United States
- F31 NS046293/NS/NINDS NIH HHS/United States
- P30 EY008126/EY/NEI NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- P60 DK20593/DK/NIDDK NIH HHS/United States
- HD15052/HD/NICHD NIH HHS/United States
- P30 DK58404/DK/NIDDK NIH HHS/United States
- R01 NS26115/NS/NINDS NIH HHS/United States
- P01 DK058212/DK/NIDDK NIH HHS/United States
- P01 HL6744/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

