A major genetic locus controlling natural Plasmodium falciparum infection is shared by East and West African Anopheles gambiae
- PMID: 17612409
- PMCID: PMC1936428
- DOI: 10.1186/1475-2875-6-87
A major genetic locus controlling natural Plasmodium falciparum infection is shared by East and West African Anopheles gambiae
Abstract
Background: Genetic linkage mapping identified a region of chromosome 2L in the Anopheles gambiae genome that exerts major control over natural infection by Plasmodium falciparum. This 2L Plasmodium-resistance interval was mapped in mosquitoes from a natural population in Mali, West Africa, and controls the numbers of P. falciparum oocysts that develop on the vector midgut. An important question is whether genetic variation with respect to Plasmodium-resistance exists across Africa, and if so whether the same or multiple geographically distinct resistance mechanisms are responsible for the trait.
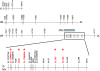
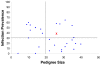
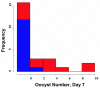
Methods: To identify P falciparum resistance loci in pedigrees generated and infected in Kenya, East Africa, 28 microsatellite loci were typed across the mosquito genome. Genetic linkage mapping was used to detect significant linkage between genotype and numbers of midgut oocysts surviving to 7-8 days post-infection.
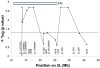
Results: A major malaria-control locus was identified on chromosome 2L in East African mosquitoes, in the same apparent position originally identified from the West African population. Presence of this resistance locus explains 75% of parasite free mosquitoes. The Kenyan resistance locus is named EA_Pfin1 (East Africa_ Plasmodium falciparum Infection Intensity).
Conclusion: Detection of a malaria-control locus at the same chromosomal location in both East and West African mosquitoes indicates that, to the level of genetic resolution of the analysis, the same mechanism of Plasmodium-resistance, or a mechanism controlled by the same genomic region, is found across Africa, and thus probably operates in A. gambiae throughout its entire range.
Figures
References
-
- World Health Organization http://www.who.int/tdr/diseases/malaria
-
- Niare O, Markianos K, Volz J, Oduol F, Toure A, Bagayoko M, Sangare D, Traore SF, Wang R, Blass C, Dolo G, Bouare M, Kafatos FC, Kruglyak L, Toure YT, Vernick KD. Genetic loci affecting resistance to human malaria parasites in a West African mosquito vector population. Science. 2002;298:213–216. doi: 10.1126/science.1073420. - DOI - PubMed
-
- Riehle MM, Markianos K, Niare O, Xu J, Li J, Toure AM, Podiougou B, Oduol F, Diawara S, Diallo M, Coulibaly B, Ouatara A, Kruglyak L, Traore SF, Vernick KD. Natural malaria infection in Anopheles gambiae is regulated by a single genomic control region. Science. 2006;312:577–579. doi: 10.1126/science.1124153. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

