Genetic diversity and population structure of wild olives from the North-Western Mediterranean assessed by SSR markers
- PMID: 17613587
- PMCID: PMC2533604
- DOI: 10.1093/aob/mcm132
Genetic diversity and population structure of wild olives from the North-Western Mediterranean assessed by SSR markers
Abstract
Background and aims: This study examines the pattern of genetic variability and genetic relationships of wild olive (Olea europaea subsp. europaea var. sylvestris) populations in the north-western Mediterranean. Recent bottleneck events are also assessed and an investigation is made of the underlying population structure of the wild olive populations.
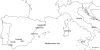
Methods: The genetic variation within and between 11 wild olive populations (171 individuals) was analysed with eight microsatellite markers. Conventional and Bayesian-based analyses were applied to infer genetic structure and define the number of gene pools in wild olive populations.
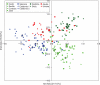
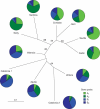
Key results: Bayesian model-based clustering identified four gene pools, which was in overall concordance with the Factorial Correspondence Analysis and Fitch-Margoliash tree. Two gene pools were predominantly found in southern Spain and Italian islands, respectively, in samples gathered from undisturbed forests of the typical Mediterranean climate. The other two gene pools were mostly detected in the north-eastern regions of Spain and in continental Italy and belong to the transition region between the temperate and Mediterranean climate zones.
Conclusions: On the basis of these results, it can be assumed that the population structure of wild olives from the north-western Mediterranean partially reflects the evolutionary history of these populations, although hybridization between true oleasters and cultivated varieties in areas of close contact between the two forms must be assumed as well. The study indicates a degree of admixture in all the populations, and suggests some caution regarding genetic differentiation at the population level, making it difficult to identify clear-cut genetic boundaries between candidate areas containing either genuinely wild or feral germplasm.
Figures
Similar articles
-
Genetic structure of wild and cultivated olives in the central Mediterranean basin.Ann Bot. 2006 Nov;98(5):935-42. doi: 10.1093/aob/mcl178. Epub 2006 Aug 25. Ann Bot. 2006. PMID: 16935868 Free PMC article.
-
Olive domestication and diversification in the Mediterranean Basin.New Phytol. 2015 Apr;206(1):436-447. doi: 10.1111/nph.13181. Epub 2014 Nov 24. New Phytol. 2015. PMID: 25420413
-
Allozyme variation of oleaster populations (wild olive tree) (Olea europaea L.) in the Mediterranean Basin.Heredity (Edinb). 2004 Apr;92(4):343-51. doi: 10.1038/sj.hdy.6800430. Heredity (Edinb). 2004. PMID: 14985782
-
How to Choose a Good Marker to Analyze the Olive Germplasm (Olea europaea L.) and Derived Products.Genes (Basel). 2021 Sep 23;12(10):1474. doi: 10.3390/genes12101474. Genes (Basel). 2021. PMID: 34680869 Free PMC article. Review.
-
Molecular studies in olive (Olea europaea L.): overview on DNA markers applications and recent advances in genome analysis.Plant Cell Rep. 2011 Apr;30(4):449-62. doi: 10.1007/s00299-010-0991-9. Epub 2011 Jan 7. Plant Cell Rep. 2011. PMID: 21212959 Review.
Cited by
-
Usefulness of a New Large Set of High Throughput EST-SNP Markers as a Tool for Olive Germplasm Collection Management.Front Plant Sci. 2018 Sep 21;9:1320. doi: 10.3389/fpls.2018.01320. eCollection 2018. Front Plant Sci. 2018. PMID: 30298075 Free PMC article.
-
Genetic Structure and Core Collection of Olive Germplasm from Albania Revealed by Microsatellite Markers.Genes (Basel). 2021 Feb 10;12(2):256. doi: 10.3390/genes12020256. Genes (Basel). 2021. PMID: 33578843 Free PMC article.
-
Conservation of Native Wild Ivory-White Olives from the MEDES Islands Natural Reserve to Maintain Virgin Olive Oil Diversity.Antioxidants (Basel). 2020 Oct 17;9(10):1009. doi: 10.3390/antiox9101009. Antioxidants (Basel). 2020. PMID: 33080812 Free PMC article.
-
Diversity of root-knot nematodes of the genus Meloidogyne Göeldi, 1892 (Nematoda: Meloidogynidae) associated with olive plants and environmental cues regarding their distribution in southern Spain.PLoS One. 2018 Jun 20;13(6):e0198236. doi: 10.1371/journal.pone.0198236. eCollection 2018. PLoS One. 2018. PMID: 29924813 Free PMC article.
-
Applications of Microsatellite Markers for the Characterization of Olive Genetic Resources of Tunisia.Genes (Basel). 2021 Feb 18;12(2):286. doi: 10.3390/genes12020286. Genes (Basel). 2021. PMID: 33670559 Free PMC article. Review.
References
-
- Alcantara JM, Rey PJ. Conflicting selection pressures on seed size: evolutionary ecology of fruit size in a bird-dispersed tree. Olea europaea. Journal of Evolutionary Biology. 2003;16:1168–1176. - PubMed
-
- Angiolillo A, Mencuccini M, Baldoni L. Olive genetic diversity assessed using amplified fragment length polymorphisms. Theoretical and Applied Genetics. 1999;98:411–421.
-
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX 4·05, logiciel sous Windows TM pour la génétique des populations. Montpellier, France: Université de Montpellier II; 2004. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000.
-
- Besnard G, Bervillé A. Multiple origins for Mediterranean olive (Olea europaea L-ssp europaea) based upon mitochondrial DNA polymorphisms. Comptes Rendus de l'Academie des Sciences, Sciences de la Vie – Life Sciences. 2000;323:173–181. - PubMed

