Precisely timed spatiotemporal patterns of neural activity in dissociated cortical cultures
- PMID: 17614210
- PMCID: PMC2096414
- DOI: 10.1016/j.neuroscience.2007.05.025
Precisely timed spatiotemporal patterns of neural activity in dissociated cortical cultures
Abstract
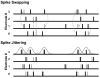
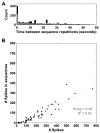
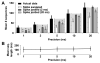
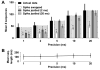
Recurring patterns of neural activity, a potential substrate of both information transfer and transformation in cortical networks, have been observed in the intact brain and in brain slices. Do these patterns require the inherent cortical microcircuitry of such preparations or are they a general property of self-organizing neuronal networks? In networks of dissociated cortical neurons from rats--which lack evidence of the intact brain's intrinsic cortical architecture--we have observed a robust set of spontaneously repeating spatiotemporal patterns of neural activity, using a template-matching algorithm that has been successful both in vivo and in brain slices. The observed patterns in cultured monolayer networks are stable over minutes of extracellular recording, occur throughout the culture's development, and are temporally precise within milliseconds. The identification of these patterns in dissociated cultures opens a powerful methodological avenue for the study of such patterns, and their persistence despite the topological and morphological rearrangements of cellular dissociation is further evidence that precisely timed patterns are a universal emergent feature of self-organizing neuronal networks.
Figures

Similar articles
-
Neuronal avalanches are diverse and precise activity patterns that are stable for many hours in cortical slice cultures.J Neurosci. 2004 Jun 2;24(22):5216-29. doi: 10.1523/JNEUROSCI.0540-04.2004. J Neurosci. 2004. PMID: 15175392 Free PMC article.
-
Understanding the temporal evolution of neuronal connectivity in cultured networks using statistical analysis.BMC Neurosci. 2014 Jan 21;15:17. doi: 10.1186/1471-2202-15-17. BMC Neurosci. 2014. PMID: 24443925 Free PMC article.
-
Deterministic neural dynamics transmitted through neural networks.Neural Netw. 2008 Aug;21(6):799-809. doi: 10.1016/j.neunet.2008.06.014. Epub 2008 Jun 28. Neural Netw. 2008. PMID: 18675536
-
Sparse and powerful cortical spikes.Curr Opin Neurobiol. 2010 Jun;20(3):306-12. doi: 10.1016/j.conb.2010.03.006. Epub 2010 Apr 17. Curr Opin Neurobiol. 2010. PMID: 20400290 Review.
-
Synchrony is stubborn in feedforward cortical networks.Nat Neurosci. 2003 Jun;6(6):543-4. doi: 10.1038/nn0603-543. Nat Neurosci. 2003. PMID: 12771956 Review. No abstract available.
Cited by
-
Detection of M-sequences from spike sequence in neuronal networks.Comput Intell Neurosci. 2012;2012:862579. doi: 10.1155/2012/862579. Epub 2012 Jul 18. Comput Intell Neurosci. 2012. PMID: 22851966 Free PMC article.
-
Repertoires of Spike Avalanches Are Modulated by Behavior and Novelty.Front Neural Circuits. 2016 Mar 22;10:16. doi: 10.3389/fncir.2016.00016. eCollection 2016. Front Neural Circuits. 2016. PMID: 27047341 Free PMC article.
-
Selectivity of stimulus induced responses in cultured hippocampal networks on microelectrode arrays.Cogn Neurodyn. 2016 Aug;10(4):287-99. doi: 10.1007/s11571-016-9380-6. Epub 2016 Feb 22. Cogn Neurodyn. 2016. PMID: 27468317 Free PMC article.
-
Modeling the formation process of grouping stimuli sets through cortical columns and microcircuits to feature neurons.Comput Intell Neurosci. 2013;2013:290358. doi: 10.1155/2013/290358. Epub 2013 Nov 28. Comput Intell Neurosci. 2013. PMID: 24369455 Free PMC article. Review.
-
Functional clustering algorithm for the analysis of dynamic network data.Phys Rev E Stat Nonlin Soft Matter Phys. 2009 May;79(5 Pt 2):056104. doi: 10.1103/PhysRevE.79.056104. Epub 2009 May 7. Phys Rev E Stat Nonlin Soft Matter Phys. 2009. PMID: 19518518 Free PMC article.
References
-
- Abeles M. Corticonics: neural circuits of the cerebral cortex. New York, NY: Cambridge University Press; 1991.
-
- Abeles M, Bergman H, Margalit E, Vaadia E. Spatiotemporal firing patterns in the frontal-cortex of behaving monkeys. J Neurophysiol. 1993;70:1629–1638. - PubMed
-
- Abeles M, Gerstein GL. Detecting spatiotemporal firing patterns among simultaneously recorded single neurons. J Neurophysiol. 1988;60:909–924. - PubMed
-
- Aertsen A, Diesmann M, Gewaltig MO. Propagation of synchronous spiking activity in feedforward neural networks. J Physiol Paris. 1996;90:243–247. - PubMed
-
- Bak P. How nature works: the science of self-organized criticality. New York, NY, USA: Copernicus; 1996.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

