Serine phosphorylation of the integrin beta4 subunit is necessary for epidermal growth factor receptor induced hemidesmosome disruption
- PMID: 17615294
- PMCID: PMC1951768
- DOI: 10.1091/mbc.e07-04-0306
Serine phosphorylation of the integrin beta4 subunit is necessary for epidermal growth factor receptor induced hemidesmosome disruption
Abstract
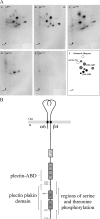
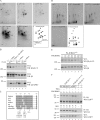
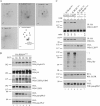
Hemidesmosomes (HDs) are multiprotein adhesion complexes that promote attachment of epithelial cells to the basement membrane. The binding of alpha6beta4 to plectin plays a central role in their assembly. We have defined three regions on beta4 that together harbor all the serine and threonine phosphorylation sites and show that three serines (S1356, S1360, and S1364), previously implicated in HD regulation, prevent the interaction of beta4 with the plectin actin-binding domain when phosphorylated. We have also established that epidermal growth factor receptor activation, which is known to function upstream of HD disassembly, results in the phosphorylation of only one or more of these three residues and the partial disassembly of HDs in keratinocytes. Additionally, we show that S1360 and S1364 of beta4 are the only residues phosphorylated by PKC and PKA in cells, respectively. Taken together, our studies indicate that multiple kinases act in concert to breakdown the structural integrity of HDs in keratinocytes, which is primarily achieved through the phosphorylation of S1356, S1360, and S1364 on the beta4 subunit.
Figures


Similar articles
-
Phosphorylation of threonine 1736 in the C-terminal tail of integrin β4 contributes to hemidesmosome disassembly.Mol Biol Cell. 2012 Apr;23(8):1475-85. doi: 10.1091/mbc.E11-11-0957. Epub 2012 Feb 22. Mol Biol Cell. 2012. PMID: 22357621 Free PMC article.
-
PKD2 and RSK1 Regulate Integrin β4 Phosphorylation at Threonine 1736.PLoS One. 2015 Nov 18;10(11):e0143357. doi: 10.1371/journal.pone.0143357. eCollection 2015. PLoS One. 2015. PMID: 26580203 Free PMC article.
-
EGF-induced MAPK signaling inhibits hemidesmosome formation through phosphorylation of the integrin {beta}4.J Biol Chem. 2010 Nov 26;285(48):37650-62. doi: 10.1074/jbc.M110.138818. Epub 2010 Sep 24. J Biol Chem. 2010. PMID: 20870721 Free PMC article.
-
Regulation of hemidesmosome disassembly by growth factor receptors.Curr Opin Cell Biol. 2008 Oct;20(5):589-96. doi: 10.1016/j.ceb.2008.05.001. Epub 2008 Jun 24. Curr Opin Cell Biol. 2008. PMID: 18583123 Review.
-
Current insights into the formation and breakdown of hemidesmosomes.Trends Cell Biol. 2006 Jul;16(7):376-83. doi: 10.1016/j.tcb.2006.05.004. Epub 2006 Jun 6. Trends Cell Biol. 2006. PMID: 16757171 Review.
Cited by
-
Syndecans and Their Synstatins: Targeting an Organizer of Receptor Tyrosine Kinase Signaling at the Cell-Matrix Interface.Front Oncol. 2021 Oct 27;11:775349. doi: 10.3389/fonc.2021.775349. eCollection 2021. Front Oncol. 2021. PMID: 34778093 Free PMC article. Review.
-
Tension in Cancer.Int J Mol Sci. 2016 Nov 16;17(11):1910. doi: 10.3390/ijms17111910. Int J Mol Sci. 2016. PMID: 27854331 Free PMC article. Review.
-
Therapeutic IMC-C225 antibody inhibits breast cancer cell invasiveness via Vav2-dependent activation of RhoA GTPase.Clin Cancer Res. 2008 Oct 1;14(19):6161-70. doi: 10.1158/1078-0432.CCR-07-5288. Clin Cancer Res. 2008. PMID: 18829495 Free PMC article.
-
The RON-receptor regulates pancreatic cancer cell migration through phosphorylation-dependent breakdown of the hemidesmosome.Int J Cancer. 2012 Oct 15;131(8):1744-54. doi: 10.1002/ijc.27447. Epub 2012 Mar 8. Int J Cancer. 2012. PMID: 22275185 Free PMC article.
-
Tetraspanin CD151 plays a key role in skin squamous cell carcinoma.Oncogene. 2013 Apr 4;32(14):1772-83. doi: 10.1038/onc.2012.205. Epub 2012 Jul 23. Oncogene. 2013. PMID: 22824799 Free PMC article.
References
-
- Boyle W. J., van der Geer P., Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–148. - PubMed
-
- Geerts D., Fontao L., Nievers M. G., Schaapveld R. Q., Purkis P. E., Wheeler G. N., Lane E. B., Leigh I. M., Sonnenberg A. Binding of integrin α6β4 to plectin prevents plectin association with F-actin but does not interfere with intermediate filament binding. J. Cell Biol. 1999;147:417–434. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

