Spatial normalization of lesioned brains: performance evaluation and impact on fMRI analyses
- PMID: 17616402
- PMCID: PMC3223520
- DOI: 10.1016/j.neuroimage.2007.04.065
Spatial normalization of lesioned brains: performance evaluation and impact on fMRI analyses
Abstract
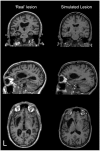
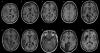
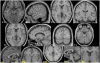
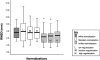
A key component of group analyses of neuroimaging data is precise and valid spatial normalization (i.e., inter-subject image registration). When patients have structural brain lesions, such as a stroke, this process can be confounded by the lack of correspondence between the subject and standardized template images. Current procedures for dealing with this problem include regularizing the estimate of warping parameters used to match lesioned brains to the template, or "cost function masking"; both these solutions have significant drawbacks. We report three experiments that identify the best spatial normalization for structurally damaged brains and establish whether differences among normalizations have a significant effect on inferences about functional activations. Our novel protocols evaluate the effects of different normalization solutions and can be applied easily to any neuroimaging study. This has important implications for users of both structural and functional imaging techniques in the study of patients with structural brain damage.
Figures
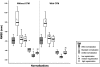

References
-
- Ardekani B., Bachman A., Strother S.C., Fujibayashi Y., Yonekura Y. Impact of inter-subjects image registration on group analysis of fMRI data. Int. Congr. Ser. 2004;1265:49–59. Ref Type: Journal (Full)
-
- Ashburner J., Friston K. Multimodal image coregistration and partitioning—A unified framework. NeuroImage. 1997;6:209–217. - PubMed
-
- Ashburner J., Friston K.J. Unified segmentation. NeuroImage. 2005;26:839–851. - PubMed
-
- Ashburner J., Neelin P., Collins D.L., Evans A., Friston K. Incorporating prior knowledge into image registration. NeuroImage. 1997;6:344–352. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

