Cellular cofactors affecting hepatitis C virus infection and replication
- PMID: 17616579
- PMCID: PMC1937561
- DOI: 10.1073/pnas.0704894104
Cellular cofactors affecting hepatitis C virus infection and replication
Abstract
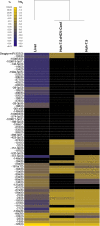
Recently identified hepatitis C virus (HCV) isolates that are infectious in cell culture provide a genetic system to evaluate the significance of virus-host interactions for HCV replication. We have completed a systematic RNAi screen wherein siRNAs were designed that target 62 host genes encoding proteins that physically interact with HCV RNA or proteins or belong to cellular pathways thought to modulate HCV infection. This includes 10 host proteins that we identify in this study to bind HCV NS5A. siRNAs that target 26 of these host genes alter infectious HCV production >3-fold. Included in this set of 26 were siRNAs that target Dicer, a principal component of the RNAi silencing pathway. Contrary to the hypothesis that RNAi is an antiviral pathway in mammals, as has been reported for subgenomic HCV replicons, siRNAs that target Dicer inhibited HCV replication. Furthermore, siRNAs that target several other components of the RNAi pathway also inhibit HCV replication. MicroRNA profiling of human liver, human hepatoma Huh-7.5 cells, and Huh-7.5 cells that harbor replicating HCV demonstrated that miR-122 is the predominant microRNA in each environment. miR-122 has been previously implicated in positively regulating the replication of HCV genotype 1 replicons. We find that 2'-O-methyl antisense oligonucleotide depletion of miR-122 also inhibits HCV genotype 2a replication and infectious virus production. Our data define 26 host genes that modulate HCV infection and indicate that the requirement for functional RNAi for HCV replication is dominant over any antiviral activity this pathway may exert against HCV.
Conflict of interest statement
Conflict of interest statement: C.M.R. has equity in Apath, LLC, which has a commercial license for the Huh-7.5 cell line and certain HCVcc derivatives.
Figures

Comment in
-
Hepatitis C virus host cell interactions uncovered.Proc Natl Acad Sci U S A. 2007 Aug 14;104(33):13215-6. doi: 10.1073/pnas.0705890104. Epub 2007 Aug 7. Proc Natl Acad Sci U S A. 2007. PMID: 17684095 Free PMC article. No abstract available.
-
RNAi: a powerful tool to unravel hepatitis C virus-host interactions within the infectious life cycle.J Hepatol. 2008 Mar;48(3):523-5. doi: 10.1016/j.jhep.2007.12.007. Epub 2007 Dec 31. J Hepatol. 2008. PMID: 18192054
References
-
- Wkly Epidemiol Rec. 1997;72:341–344. Anonymous. - PubMed
-
- Tellinghuisen TL, Rice CM. Curr Opin Microbiol. 2002;5:419–427. - PubMed
-
- Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Science. 1999;285:110–113. - PubMed
-
- Blight KJ, Kolykhalov AA, Rice CM. Science. 2000;290:1972–1974. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA085883/CA/NCI NIH HHS/United States
- CA 107092/CA/NCI NIH HHS/United States
- P30 DK 42086/DK/NIDDK NIH HHS/United States
- CA 57973/CA/NCI NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- AI 40034/AI/NIAID NIH HHS/United States
- F32 AI051820/AI/NIAID NIH HHS/United States
- 5F32 AI 51820-03/AI/NIAID NIH HHS/United States
- N01 AI040034/AI/NIAID NIH HHS/United States
- P30 DK042086/DK/NIDDK NIH HHS/United States
- CA 85883/CA/NCI NIH HHS/United States
- K01 CA107092/CA/NCI NIH HHS/United States
- R01 CA057973/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

