Regulation of gbpC expression in Streptococcus mutans
- PMID: 17616585
- PMCID: PMC2045159
- DOI: 10.1128/JB.00825-07
Regulation of gbpC expression in Streptococcus mutans
Abstract
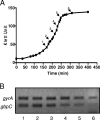
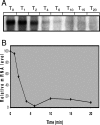
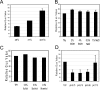
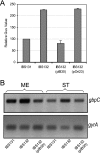
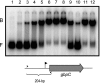
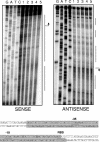
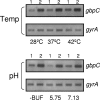
Streptococcus mutans, the principal causative agent of dental caries, produces four glucan-binding proteins (Gbp) that play major roles in bacterial adherence and pathogenesis. One of these proteins, GbpC, is an important cell surface protein involved in biofilm formation. GbpC is also important for cariogenesis, bacteremia, and infective endocarditis. In this study, we examined the regulation of gbpC expression in S. mutans strain UA159. We found that gbpC expression attains the maximum level at mid-exponential growth phase, and the half-life of the transcript is less than 2 min. Expression from PgbpC was measured using a PgbpC-gusA transcriptional fusion reporter and was analyzed under various stress conditions, including thermal, osmotic, and acid stresses. Expression of gbpC is induced under conditions of thermal stress but is repressed during growth at low pH, whereas osmotic stress had no effect on expression from PgbpC. The results from the expression analyses were further confirmed using semiquantitative reverse transcription-PCR analysis. Our results also reveal that CovR, a global response regulator in many Streptococcus spp., represses gbpC expression at the transcriptional level. We demonstrated that purified CovR protein binds directly to the promoter region of PgbpC to repress gbpC expression. Using a DNase I protection assay, we showed that CovR binds to DNA sequences surrounding PgbpC from bases -68 to 28 (where base 1 is the start of transcription). In summary, our results indicate that various stress conditions modulate the expression of gbpC and that CovR negatively regulates the expression of the gbpC gene by directly binding to the promoter region.
Figures

Similar articles
-
LiaS regulates virulence factor expression in Streptococcus mutans.Infect Immun. 2008 Jul;76(7):3093-9. doi: 10.1128/IAI.01627-07. Epub 2008 May 5. Infect Immun. 2008. PMID: 18458070 Free PMC article.
-
[Location of GbpC protein in Streptococcus mutans UA159].Zhonghua Kou Qiang Yi Xue Za Zhi. 2007 Jun;42(6):349-52. Zhonghua Kou Qiang Yi Xue Za Zhi. 2007. PMID: 17888251 Chinese.
-
Modulation of covR expression in Streptococcus mutans UA159.J Bacteriol. 2008 Jul;190(13):4478-88. doi: 10.1128/JB.01961-07. Epub 2008 May 9. J Bacteriol. 2008. PMID: 18469111 Free PMC article.
-
Role of the Streptococcus mutans irvA gene in GbpC-independent, dextran-dependent aggregation and biofilm formation.Appl Environ Microbiol. 2009 Nov;75(22):7037-43. doi: 10.1128/AEM.01015-09. Epub 2009 Sep 25. Appl Environ Microbiol. 2009. PMID: 19783751 Free PMC article.
-
Identification of a glucan-binding protein C gene homologue in Streptococcus macacae.Oral Microbiol Immunol. 2006 Feb;21(1):32-41. doi: 10.1111/j.1399-302X.2005.00251.x. Oral Microbiol Immunol. 2006. PMID: 16390339
Cited by
-
Pluronics-Formulated Farnesol Promotes Efficient Killing and Demonstrates Novel Interactions with Streptococcus mutans Biofilms.PLoS One. 2015 Jul 29;10(7):e0133886. doi: 10.1371/journal.pone.0133886. eCollection 2015. PLoS One. 2015. PMID: 26222384 Free PMC article.
-
A model of efficiency: stress tolerance by Streptococcus mutans.Microbiology (Reading). 2008 Nov;154(Pt 11):3247-3255. doi: 10.1099/mic.0.2008/023770-0. Microbiology (Reading). 2008. PMID: 18957579 Free PMC article. Review.
-
Amyloid Fibrils Produced by Streptococcus sanguinis Contribute to Biofilm Formation and Immune Evasion.Int J Mol Sci. 2023 Oct 28;24(21):15686. doi: 10.3390/ijms242115686. Int J Mol Sci. 2023. PMID: 37958670 Free PMC article.
-
Role of CovR phosphorylation in gene transcription in Streptococcus mutans.Microbiology (Reading). 2018 Apr;164(4):704-715. doi: 10.1099/mic.0.000641. Epub 2018 Mar 5. Microbiology (Reading). 2018. PMID: 29504927 Free PMC article.
-
The Streptococcus mutans IrvR repressor is a CI-like regulator that functions through autocleavage and Clp-dependent proteolysis.J Bacteriol. 2010 Mar;192(6):1586-95. doi: 10.1128/JB.01261-09. Epub 2009 Dec 28. J Bacteriol. 2010. PMID: 20038591 Free PMC article.
References
-
- Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 99:14434-14439. - PMC - PubMed
-
- Banas, J. A., and M. M. Vickerman. 2003. Glucan-binding proteins of the oral streptococci. Crit. Rev. Oral Biol. Med. 14:89-99. - PubMed
-
- Browngardt, C. M., Z. T. Wen, and R. A. Burne. 2004. RegM is required for optimal fructosyltransferase and glucosyltransferase gene expression in Streptococcus mutans. FEMS Microbiol. Lett. 240:75-79. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

