Molecular characterization of a novel ortho-nitrophenol catabolic gene cluster in Alcaligenes sp. strain NyZ215
- PMID: 17616586
- PMCID: PMC2045184
- DOI: 10.1128/JB.00654-07
Molecular characterization of a novel ortho-nitrophenol catabolic gene cluster in Alcaligenes sp. strain NyZ215
Abstract
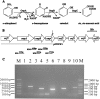
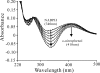
Alcaligenes sp. strain NyZ215 was isolated for its ability to grow on ortho-nitrophenol (ONP) as the sole source of carbon, nitrogen, and energy and was shown to degrade ONP via a catechol ortho-cleavage pathway. A 10,152-bp DNA fragment extending from a conserved region of the catechol 1,2-dioxygenase gene was obtained by genome walking. Of seven complete open reading frames deduced from this fragment, three (onpABC) have been shown to encode the enzymes involved in the initial reactions of ONP catabolism in this strain. OnpA, which shares 26% identity with salicylate 1-monooxygenase of Pseudomonas stutzeri AN10, is an ONP 2-monooxygenase (EC 1.14.13.31) which converts ONP to catechol in the presence of NADPH, with concomitant nitrite release. OnpC is a catechol 1,2-dioxygenase catalyzing the oxidation of catechol to cis,cis-muconic acid. OnpB exhibits 54% identity with the reductase subunit of vanillate O-demethylase in Pseudomonas fluorescens BF13. OnpAB (but not OnpA alone) conferred on the catechol utilizer Pseudomonas putida PaW340 the ability to grow on ONP. This suggests that OnpB may also be involved in ONP degradation in vivo as an o-benzoquinone reductase converting o-benzoquinone to catechol. This is analogous to the reduction of tetrachlorobenzoquinone to tetrachlorohydroquinone by a tetrachlorobenzoquinone reductase (PcpD, 38% identity with OnpB) in the pentachlorophenol degrader Sphingobium chlorophenolicum ATCC 39723.
Figures

Similar articles
-
Identification and characterization of catabolic para-nitrophenol 4-monooxygenase and para-benzoquinone reductase from Pseudomonas sp. strain WBC-3.J Bacteriol. 2009 Apr;191(8):2703-10. doi: 10.1128/JB.01566-08. Epub 2009 Feb 13. J Bacteriol. 2009. PMID: 19218392 Free PMC article.
-
Characterization of a para-nitrophenol catabolic cluster in Pseudomonas sp. strain NyZ402 and construction of an engineered strain capable of simultaneously mineralizing both para- and ortho-nitrophenols.Biodegradation. 2010 Jul;21(4):575-84. doi: 10.1007/s10532-009-9325-4. Epub 2010 Jan 5. Biodegradation. 2010. PMID: 20049512
-
Construction of an engineered strain free of antibiotic resistance gene markers for simultaneous mineralization of methyl parathion and ortho-nitrophenol.Appl Microbiol Biotechnol. 2010 Jun;87(1):281-7. doi: 10.1007/s00253-010-2457-y. Epub 2010 Feb 16. Appl Microbiol Biotechnol. 2010. PMID: 20157808
-
Influence of para-substituents on the oxidative metabolism of o-nitrophenols by Pseudomonas putida B2.Appl Environ Microbiol. 1986 Aug;52(2):334-9. doi: 10.1128/aem.52.2.334-339.1986. Appl Environ Microbiol. 1986. PMID: 3752997 Free PMC article.
-
Cloning of a gene cluster involved in the catabolism of p-nitrophenol by Arthrobacter sp. strain JS443 and characterization of the p-nitrophenol monooxygenase.J Bacteriol. 2007 Nov;189(21):7563-72. doi: 10.1128/JB.01849-06. Epub 2007 Aug 24. J Bacteriol. 2007. PMID: 17720792 Free PMC article.
Cited by
-
Nitroaromatic compounds, from synthesis to biodegradation.Microbiol Mol Biol Rev. 2010 Jun;74(2):250-72. doi: 10.1128/MMBR.00006-10. Microbiol Mol Biol Rev. 2010. PMID: 20508249 Free PMC article. Review.
-
A radical intermediate in the conversion of pentachlorophenol to tetrachlorohydroquinone by Sphingobium chlorophenolicum.Biochemistry. 2014 Oct 21;53(41):6539-49. doi: 10.1021/bi5010427. Epub 2014 Oct 6. Biochemistry. 2014. PMID: 25238136 Free PMC article.
-
Identification and characterization of catabolic para-nitrophenol 4-monooxygenase and para-benzoquinone reductase from Pseudomonas sp. strain WBC-3.J Bacteriol. 2009 Apr;191(8):2703-10. doi: 10.1128/JB.01566-08. Epub 2009 Feb 13. J Bacteriol. 2009. PMID: 19218392 Free PMC article.
-
Genetic and Biochemical Characterization of 2-Chloro-5-Nitrophenol Degradation in a Newly Isolated Bacterium, Cupriavidus sp. Strain CNP-8.Front Microbiol. 2017 Sep 13;8:1778. doi: 10.3389/fmicb.2017.01778. eCollection 2017. Front Microbiol. 2017. PMID: 28959252 Free PMC article.
-
MhbT is a specific transporter for 3-hydroxybenzoate uptake by Gram-negative bacteria.Appl Environ Microbiol. 2012 Sep;78(17):6113-20. doi: 10.1128/AEM.01511-12. Epub 2012 Jun 22. Appl Environ Microbiol. 2012. PMID: 22729544 Free PMC article.
References
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
-
- de Lorenzo, V., L. Eltis, B. Kessler, and K. N. Timmis. 1993. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene 123:17-24. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

