The serine protease HhoA from Synechocystis sp. strain PCC 6803: substrate specificity and formation of a hexameric complex are regulated by the PDZ domain
- PMID: 17616590
- PMCID: PMC2045181
- DOI: 10.1128/JB.00883-07
The serine protease HhoA from Synechocystis sp. strain PCC 6803: substrate specificity and formation of a hexameric complex are regulated by the PDZ domain
Abstract
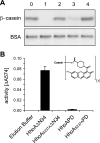
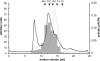
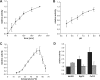
Enzymes of the ATP-independent Deg serine endopeptidase family are very flexible with regard to their substrate specificity. Some family members cleave only one substrate, while others act as general proteases on unfolded substrates. The proteolytic activity of Deg proteases is regulated by PDZ protein interaction domains. Here we characterized the HhoA protease from Synechocystis sp. strain PCC 6803 in vitro using several recombinant protein constructs. The proteolytic activity of HhoA was found to increase with temperature and basic pH and was stimulated by the addition of Mg(2+) or Ca(2+). We found that the single PDZ domain of HhoA played a critical role in regulating protease activity and in the assembly of a hexameric complex. Deletion of the PDZ domain strongly reduced proteolysis of a sterically challenging resorufin-labeled casein substrate, but unlabeled beta-casein was still degraded. Reconstitution of the purified HhoA with total membrane proteins isolated from Synechocystis sp. wild-type strain PCC 6803 and a DeltahhoA mutant resulted in specific degradation of selected proteins at elevated temperatures. We concluded that a single PDZ domain of HhoA plays a critical role in defining the protease activity and oligomerization state, combining the functions that are attributed to two PDZ domains in the homologous DegP protease from Escherichia coli. Based on this first enzymatic study of a Deg protease from cyanobacteria, we propose a general role for HhoA in the quality control of extracytoplasmic proteins, including membrane proteins, in Synechocystis sp. strain PCC 6803.
Figures
Similar articles
-
Proteomic approaches to identify substrates of the three Deg/HtrA proteases of the cyanobacterium Synechocystis sp. PCC 6803.Biochem J. 2015 Jun 15;468(3):373-84. doi: 10.1042/BJ20150097. Epub 2015 Apr 16. Biochem J. 2015. PMID: 25877158
-
The HhoA protease from Synechocystis sp. PCC 6803 - Novel insights into structure and activity regulation.J Struct Biol. 2017 Jun;198(3):147-153. doi: 10.1016/j.jsb.2016.12.004. Epub 2016 Dec 10. J Struct Biol. 2017. PMID: 27956128
-
Crystal structure of the zinc-bound HhoA protease from Synechocystis sp. PCC 6803.FEBS Lett. 2016 Oct;590(19):3435-3442. doi: 10.1002/1873-3468.12416. Epub 2016 Sep 23. FEBS Lett. 2016. PMID: 27616292
-
Cellular functions, mechanism of action, and regulation of FtsH protease.Annu Rev Microbiol. 2005;59:211-31. doi: 10.1146/annurev.micro.59.030804.121316. Annu Rev Microbiol. 2005. PMID: 15910274 Review.
-
Bioinformatics perspective on rhomboid intramembrane protease evolution and function.Biochim Biophys Acta. 2013 Dec;1828(12):2937-43. doi: 10.1016/j.bbamem.2013.06.031. Epub 2013 Jul 8. Biochim Biophys Acta. 2013. PMID: 23845876 Free PMC article. Review.
Cited by
-
Recombinant Deg/HtrA proteases from Synechocystis sp. PCC 6803 differ in substrate specificity, biochemical characteristics and mechanism.Biochem J. 2011 May 1;435(3):733-42. doi: 10.1042/BJ20102131. Biochem J. 2011. PMID: 21332448 Free PMC article.
-
Degradation of PsbO by the Deg protease HhoA Is thioredoxin dependent.PLoS One. 2012;7(9):e45713. doi: 10.1371/journal.pone.0045713. Epub 2012 Sep 19. PLoS One. 2012. PMID: 23029195 Free PMC article.
-
Biochemical characterization of the SPATE members EspPα and EspI.Toxins (Basel). 2014 Sep 16;6(9):2719-31. doi: 10.3390/toxins6092719. Toxins (Basel). 2014. PMID: 25229188 Free PMC article.
-
Programmed cell death in plants: A chloroplastic connection.Plant Signal Behav. 2015;10(2):e989752. doi: 10.4161/15592324.2014.989752. Plant Signal Behav. 2015. PMID: 25760871 Free PMC article. Review.
References
-
- Alba, B. M., H. J. Zhong, J. C. Pelayo, and C. A. Gross. 2001. degS (hhoB) is an essential Escherichia coli gene whose indispensable function is to provide sigma(E) activity. Mol. Microbiol. 40:1323-1333. - PubMed
-
- Barker, M., R. de Vries, J. Nield, J. Komenda, and P. J. Nixon. 2006. The deg proteases protect Synechocystis sp. PCC 6803 during heat and light stresses but are not essential for removal of damaged D1 protein during the photosystem two repair cycle. J. Biol. Chem. 281:30347-30355. - PubMed
-
- Cavard, D. 1995. Role of DegP protease on levels of various forms of colicin A lysis protein. FEMS Microbiol. Lett. 125:173-178. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

