Interactions of the antizyme AtoC with regulatory elements of the Escherichia coli atoDAEB operon
- PMID: 17616594
- PMCID: PMC1951910
- DOI: 10.1128/JB.00214-07
Interactions of the antizyme AtoC with regulatory elements of the Escherichia coli atoDAEB operon
Abstract
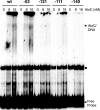
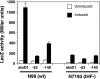
AtoC has a dual function as both an antizyme, the posttranslational inhibitor of polyamine biosynthetic enzymes, and the transcriptional regulator of genes involved in short-chain fatty acid catabolism (the atoDAEB operon). We have previously shown that AtoC is the response regulator of the AtoS-AtoC two-component signal transduction system that activates atoDAEB when Escherichia coli is exposed to acetoacetate. Here, we show that the same cis elements control both promoter inducibility and AtoC binding. Chromatin immunoprecipitation experiments confirmed the acetoacetate-inducible binding of AtoC to the predicted DNA region in vivo. DNase I protection footprinting analysis revealed that AtoC binds two 20-bp stretches, constituting an inverted palindrome, that are located at -146 to -107 relative to the transcription initiation site. Analyses of promoter mutants obtained by in vitro chemical mutagenesis of the atoDAEB promoter verified both the importance of AtoC binding for the inducibility of the promoter by acetoacetate and the sigma54 dependence of atoDAEB expression. The integration host factor was also identified as a critical component of the AtoC-mediated induction of atoDAEB.
Figures

Similar articles
-
Escherichia coli genome-wide promoter analysis: identification of additional AtoC binding target elements.BMC Genomics. 2011 May 13;12(1):238. doi: 10.1186/1471-2164-12-238. BMC Genomics. 2011. PMID: 21569465 Free PMC article.
-
Phosphorylation activity of the response regulator of the two-component signal transduction system AtoS-AtoC in E. coli.Biochim Biophys Acta. 2005 Oct 10;1725(3):257-68. doi: 10.1016/j.bbagen.2005.06.019. Biochim Biophys Acta. 2005. PMID: 16153782
-
Effect of polyamines and synthetic polyamine-analogues on the expression of antizyme (AtoC) and its regulatory genes.BMC Biochem. 2007 Jan 15;8:1. doi: 10.1186/1471-2091-8-1. BMC Biochem. 2007. PMID: 17224065 Free PMC article.
-
Activation of the AtoSC two-component system in the absence of the AtoC N-terminal receiver domain in E. coli.Amino Acids. 2011 Feb;40(2):421-30. doi: 10.1007/s00726-010-0652-x. Epub 2010 Jun 19. Amino Acids. 2011. PMID: 20563612
-
Signal transduction and adaptive regulation through bacterial two-component systems: the Escherichia coli AtoSC paradigm.Amino Acids. 2009 Sep;37(3):443-58. doi: 10.1007/s00726-009-0241-z. Epub 2009 Feb 8. Amino Acids. 2009. PMID: 19198978 Review.
Cited by
-
Genome sequence and analysis of Escherichia coli production strain LS5218.Metab Eng Commun. 2017 Nov 2;5:78-83. doi: 10.1016/j.meteno.2017.10.001. eCollection 2017 Dec. Metab Eng Commun. 2017. PMID: 29188187 Free PMC article.
-
Insights into coastal microbial antibiotic resistome through a meta-transcriptomic approach in Yucatan.Front Microbiol. 2022 Oct 17;13:972267. doi: 10.3389/fmicb.2022.972267. eCollection 2022. Front Microbiol. 2022. PMID: 36325016 Free PMC article.
-
Escherichia coli genome-wide promoter analysis: identification of additional AtoC binding target elements.BMC Genomics. 2011 May 13;12(1):238. doi: 10.1186/1471-2164-12-238. BMC Genomics. 2011. PMID: 21569465 Free PMC article.
-
A metabolic pathway for catabolizing levulinic acid in bacteria.Nat Microbiol. 2017 Dec;2(12):1624-1634. doi: 10.1038/s41564-017-0028-z. Epub 2017 Sep 25. Nat Microbiol. 2017. PMID: 28947739 Free PMC article.
-
Exploring Fatty Acid β-Oxidation Pathways in Bacteria: From General Mechanisms to DSF Signaling and Pathogenicity in Xanthomonas.Curr Microbiol. 2024 Sep 2;81(10):336. doi: 10.1007/s00284-024-03866-8. Curr Microbiol. 2024. PMID: 39223428 Review.
References
-
- Clark, D. P., and J. E. Cronan, Jr. 1996. Two-carbon compounds and fatty acids as carbon sources, p. 343-357. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, DC.
-
- Cotter, P. A., and V. J. DiRita. 2000. Bacterial virulence gene regulation: an evolutionary perspective. Annu. Rev. Microbiol. 54:519-565. - PubMed
-
- Da Re, S. S., D. Deville-Bonne, T. Tolstykh, M. Veron, and J. B. Stock. 1999. Kinetics of CheY phosphorylation by small molecule phosphodonors. FEBS Lett. 457:323-326. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

