Temporal dynamics and impact of manure storage on antibiotic resistance patterns and population structure of Escherichia coli isolates from a commercial swine farm
- PMID: 17616622
- PMCID: PMC2042059
- DOI: 10.1128/AEM.00218-07
Temporal dynamics and impact of manure storage on antibiotic resistance patterns and population structure of Escherichia coli isolates from a commercial swine farm
Abstract
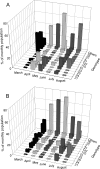
Many confined-livestock farms store their wastes for several months prior to use as a fertilizer. Storing manure for extended periods could significantly bias the composition of enteric bacterial populations subsequently released into the environment. Here, we compared populations of Escherichia coli isolated from fresh feces and from the manure-holding tank (stored manure) of a commercial swine farm, each sampled monthly for 6 months. The 4,668 confirmed E. coli isolates were evaluated for resistance to amikacin, ampicillin, cephalothin, chloramphenicol, kanamycin, nalidixic acid, streptomycin, sulfamethoxazole, tetracycline, trimethoprim, and trimethoprim plus sulfamethoxazole. A subset of 1,687 isolates was fingerprinted by repetitive extragenic palindromic PCR (rep-PCR) with the BOXA1R primer to evaluate the diversity and the population structure of the collection. The population in the stored manure was generally more diverse than that in the fresh feces. Half of the genotypes detected in the stored manure were never detected in the fresh fecal material, and only 16% were detected only in the fresh feces. But the majority of the isolates (84%) were assigned to the 34% of genotypes shared between the two environments. The structure of the E. coli population showed important monthly variations both in the extent and distribution of the diversity of the observed genotypes. The frequency of detection of resistance to specific antibiotics was not significantly different between the two collections and varied importantly between monthly samples. Resistance to multiple antibiotics was much more temporally dynamic in the fresh feces than in the stored manure. There was no relationship between the distribution of rep-PCR fingerprints and the distribution of antibiotic resistance profiles, suggesting that specific antibiotic resistance determinants were dynamically distributed within the population.
Figures
References
-
- Anonymous. 2003. Assessing microbial safety of drinking water: improving approaches and methods. OECD, WHO, Paris, France.
-
- Bischoff, K. M., D. G. White, M. E. Hume, T. L. Poole, and D. J. Nisbet. 2005. The chloramphenicol resistance gene cmlA is disseminated on transferable plasmids that confer multiple-drug resistance in swine Escherichia coli. FEMS Microbiol. Lett. 243:285-291. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical